What are the recent trends in Structural Biology and Drug Discovery? - Insights from the PSDI conference
Among the conferences I have attended, the Protein Structure Determination in Industry (PSDI) meeting is definitely one of my favorites. This is mainly due to the exceptional quality of the scientific content and the way it brings together the structural biology community (mainly from industry). The conference offers an amazing opportunity to learn about new trends in the field and discuss them with fellow scientists.
This year, the 32nd edition was held in Paris and was, as usual, wonderfully organized thanks to the scientific committee and the collaboration with the Synchrotron SOLEIL. Discngine is a constant supporter and sponsor of the event, so I had the privilege to attend as one of the sponsor delegates, alongside my colleagues.
Discngine delegation the 32nd Protein Structure Determination in Industry (PSDI) meeting in Paris, France.
This edition included industry talks across various areas of structural biology, highlighting several key advancements and emerging methods in protein structure determination (the event page is still live, and you can check the full agenda here).
There was also an engaging poster session, where I had the opportunity to present my poster on the accuracy of protein-protein interactions predictions in AF3 models of PD-1 complexes (more at the end of the article).
In this blog post I will summarize my key learnings from the event, including:
The impact of Cryo-EM structures on drug discovery
Integration of AI in drug discovery workflows
Emerging Techniques and Modalities
Discover event details and join us!
We are organizing a dedicated event focusing on the synergies of AI and experimental approaches for efficient SBDD in Cambridge, UK on March 19th!
Join us for a scientific program and networking session in a pleasant atmosphere to exchange thoughts, experiences and build connections.
Check out agenda, speakers and register here.
The impact of Cryo-EM structures on drug discovery
Needless to say, recent advancements in Cryo-Electron Microscopy (Cryo-EM) bring tremendous opportunities for drug discovery. Cryo-EM high-resolution structures have enabled the structural characterization of challenging targets, such as integral membrane proteins (IMP), complementing precedented protein structure determination methods. Throughout the conference, numerous talks highlighted the crucial contribution of Cryo-EM in determining the 3D structures of protein targets, which facilitated the application of structure-based approaches for drug discovery projects.
One notable example was the compelling presentation by Dietmar Weichert (Boehringer Ingelheim) reporting structural insights on the orphan class A G-Protein Coupled Receptor (GPCR) GPR55. He showcased how enhancements in the structure determination process by Cryo-EM have significantly supported their structure-based drug discovery (SBDD) campaigns for challenging targets. The Cryo-EM structure of GPR55 allowed detailed analysis of the GPR55-G protein complex architecture and revealed the binding mode of the native ligand, leading to a better understanding of receptor activation. This structural information guided the design of new drugs targeting GPR55. (paper under revision - Claff, T. et al. Structural basis of lipid-mediated activation of G-protein coupled receptor GPR55. Nat Comm)
Weichert also highlighted that Cryo-EM was the method of choice for solving 3D structures in their SBDD projects on difficult targets, achieving an impressive 86% success rate for IMP. This insight shows the superior capability of Cryo-EM in tackling the challenges of structural determination of IMP, which often proves resistant to other structural determination techniques. As a result, this streamlined their SBDD pipeline, reducing project completion time to an average of 16 months for new challenging targets.
Dietmar Weichert’s talk “Enabling Structure-based Drug Discovery for Membrane Proteins: Insights Into Lipid-mediated Activation of GPR55”
In addition to other success stories on Cryo-EM contributions to SBDD, an inspiring talk by James Kiefer (Genentech) illustrated how to overcome current technique limitations to further expand its application in drug discovery. A well-known limit of Cryo-EM is the size constraint on the target, with only large proteins or protein complexes (> 50 kDa) typically suitable for high-resolution determination. Kiefer reported a new method that addresses this restriction, allowing Cryo-EM to achieve high-resolution structures of small proteins (15-20 kDa).
This approach stabilizes the protein of interest by binding it with antibody fragments (Fabs) that have an exceptional rigidity (disulfide constrained Fabs; “Rigid-Fabs”) compared to other systems previously used with this purpose. The higher rigidity limits the conformational flexibility of the protein, which gains sufficient stability to allow for high-resolution structure determination by Cryo-EM.
These and many other real-life stories shared at the event exemplify how Cryo-EM is transforming drug discovery. By providing crucial molecular details on challenging targets, it ultimately accelerates the development of novel therapeutic strategies.
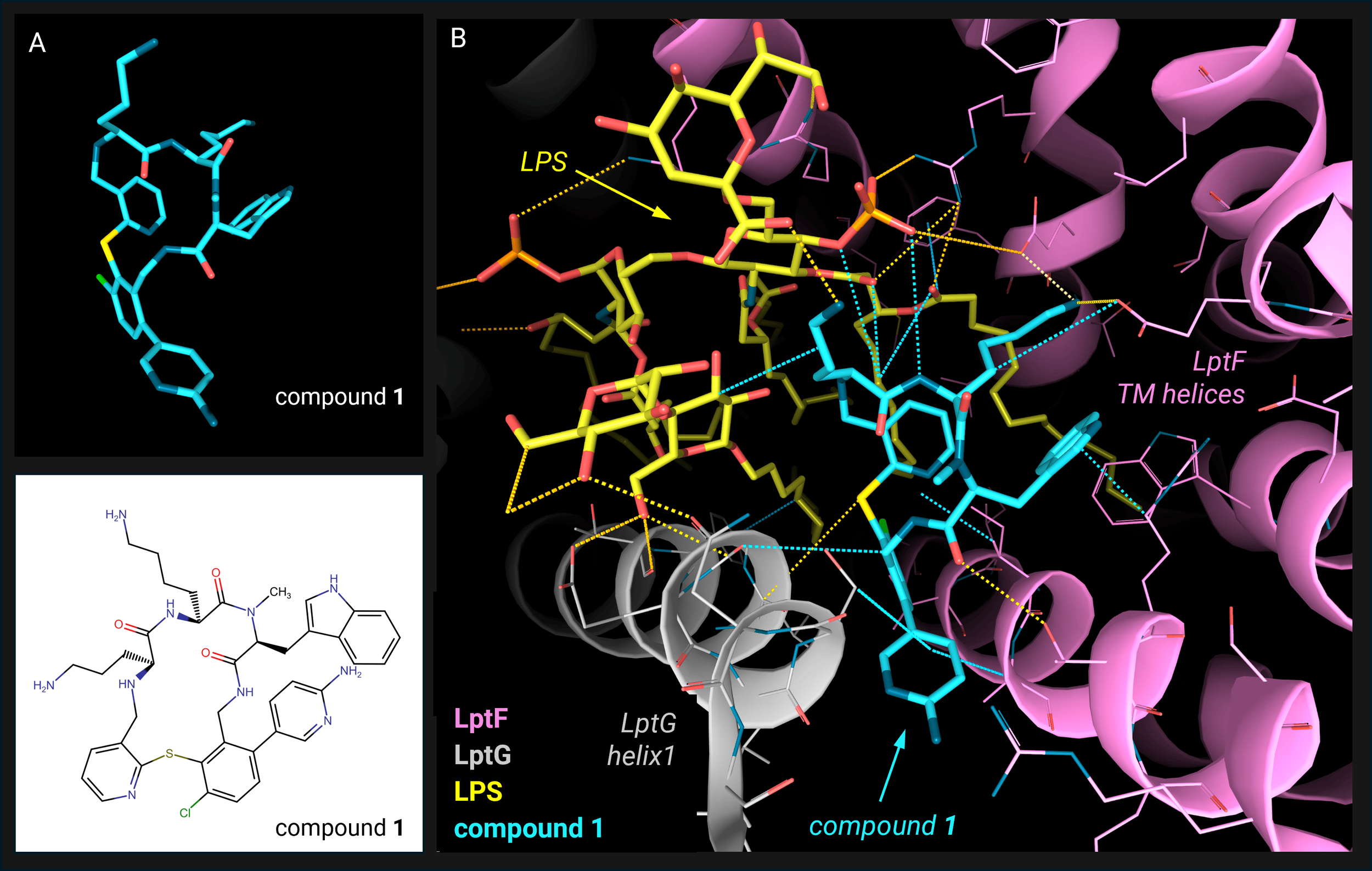
Interested in the case study on how Cryo-EM is transforming the discovery of antibiotic drugs?
Thomas Clairfeuille presented how the Roche team is fighting antibiotic resistance by designing novel, effective drugs guided by insights from high-resolution cryo-EM structures. Learn about this study in our December Protein of the Month featuring LPS transporter (Lpt).
Integration of AI in drug discovery workflows
Artificial Intelligence (AI) technologies are being increasingly adopted into drug discovery workflows, due to their ability to analyze vast datasets, identify patterns, and make predictions, thus streamlining the drug discovery process.
In the context of Structural Biology, 3D structure prediction algorithms such as AlphaFold 2 (AF2) and RoseTTA had a massive impact, as acknowledged by the 2024 Nobel Prize in Chemistry. Throughout many presentations at the PSDI conference, one could evidently notice the growing synergy between AI and traditional experimental approaches. It is clear that predictive technologies have been rapidly integrated in pharma and biotech SBDD workflows, especially for:
Construct optimization: AF2 models have facilitated the identification of domains to be structurally solved, thereby supporting construct design. This contribution is of pivotal importance, since one of the most demanding aspects of 3D protein structure determination for difficult targets is still producing a stable, well-expressed construct. AI-driven construct optimization significantly enhances the efficiency and success rate of these efforts.
Model Refinement: using AI-predicted 3D protein structures for Molecular Replacement (MR) in the refinement of experimental structures has become one of the most straightforward and successful integrations of AF2 models into drug discovery workflows, effectively addressing the “phase problem”. In fact, AF2, in most cases, provides sufficiently accurate models that can be used as a starting point for MR, reducing the need for experimental phasing. This approach is particularly valuable for diverse folds and novel targets with low homology, and it is streamlining the determination of 3D protein structures, accelerating SBDD.
Protein engineering: AI algorithms are being developed to enhance protein engineering, producing proteins with optimized properties to support SBDD. A notable example was presented by Casper Goverde (EPFL, Switzerland), who described the AI-driven production of soluble and functional membrane protein analogs that could be effectively used for drug discovery purposes. These AI models have the ability to overcome challenges typically associated with membrane proteins structural studies.
The protein in the picture on the left was part of the study presented by Casper Goverde in his talk: "Functional Design of Soluble Analogs of Integral Membrane Proteins". It is an example of an X-ray crystal structure of a soluble protein with a GPCR-like fold (GLF_32, PDB: 8OYY), produced using AI algorithms. The protein surface is color-coded by hydrophobicity score, showing that this GPCR-like protein is mostly hydrophilic (green). In contrast, a “classic” GPCR (picture on the right, PDB: 2RH1) would exhibit a highly hydrophobic (yellow-red) surface in the transmembrane domain.
In addition to the integration of AI methods in SBDD, another exciting advance, reported and discussed by various speakers and attendees, was the potential of using 3D structure prediction algorithms for elucidating previously uncharacterized functions of proteins. In a recent paper, researchers reported the use of AlphaFold-Multimer for screening predicted protein-protein interactions. This approach identified the protein Tmem81 as part of a conserved fertilization protein complex, revealing its essential role in male fertility in fish and mice. The human orthologs of the identified complex also interact in vitro, opening exciting opportunities for addressing human infertility.
Collectively, these advancements demonstrate the transformative potential of AI in structural biology and medicine, while also opening up innovative and unprecedented research fields. As brilliantly expressed in Matthias Haffke’s (Novartis) insightful presentation: “Let AI Be Your Guide”.
“Let AI be your guide to drive ideation and hypothesis generation”
Emerging Techniques and Modalities
During the conference, various innovative technologies were presented that promise to complement current structural biology techniques and broaden the contribution of structural information to new drug modalities. Among these:
Cryo-Electron Tomography (Cryo-ET): this technique provides 3D visualization of cellular structures at near-atomic resolution. Unlike traditional Cryo-EM, Cryo-ET allows the examination of proteins within their native cellular environments, providing richer and unprecedented information that cannot be obtained from single particle Cryo-EM (which works on diluted solutions of proteins).
A fascinating example was discussed by Madeleine Gilbert (University of Leeds), who presented how Cryo-ET contributed to determining the in-tissue 3D architectures of β-amyloid plaques and tau pathology in a human postmortem Alzheimer’s disease brain. This technique can provide invaluable insights into the spatial organization and functional dynamics of biomolecules, paving the way for new discoveries in structural biology and drug development.
Targeted Protein Degraders (TPD): a groundbreaking drug modality with the potential to target proteins that were previously considered “undruggable”, exploiting the cell’s natural protein degradation machinery to selectively degrade disease-causing proteins.
In his inspiring presentation, Georg Petzold (Monte Rosa Therapeutics) showed how they developed a structure-based computational approach, starting from structural considerations on the Cereblon (CRBN) ubiquitin ligase, to mine the human proteome. This strategy predicted over 1400 putative novel target proteins, of which 22 - with clinical implications - were validated, identifying a novel mechanism of substrate recognition. These findings broaden the CRBN target space and establish a foundation for targeting more “undruggable” proteins with the next-generation TPD.
RNA: targeting RNA molecules offers innovative therapeutic approaches for various diseases, but the rational, structure-based design of small molecule compounds is hindered by the limited understanding of RNA structural and dynamic properties. "Evi" Paraskevi Gkeka (Sanofi) illustrated recent advancements in the identification of small molecule binding sites in RNA conformational ensembles, using a computational technique called “SHAMAN”. This method explores the conformational landscape of RNA with all-atom molecular dynamics simulations accelerated by metadynamics.
SHAMAN successfully identified a set of known, experimentally resolved RNA-small molecule structures, and was able to rank all identified pockets. This computational pipeline sets the foundation for future design of novel RNA drugs driven by structural insights.
All these abovementioned insights are the reason this conference is one of my favorites – the selection of techniques and methodologies highlights achievements that will likely influence the efforts of structural biologists in the next years and have the potential to significantly impact SBDD projects.
Conclusions
In this short blog post, I have summarized my main takeaways from the meeting and tried to anticipate the future trends in structural biology for the coming years. Looking back at previous editions I have attended, it is impressive to witness how rapidly groundbreaking technologies such as Cryo-EM and AI algorithms for structure determination have been embraced by the industry.
Beyond the valuable insights gained from the scientific talks of this PSDI 2024, connecting with the structural biology community and exchanging ideas with scientists was truly enriching. Moreover, the evening cruise on the river Seine made the event even more memorable! I look forward to attending the next PSDI in Basel and learning about the advancements that are shaping the future of the drug discovery industry.
Case study on the accuracy of AF3 predicted models
At the PSDI 2024 scientific poster session, I have presented my findings on the ability of novel AF3 models to predict protein-protein interfaces and epitopes. You can find the full case study bellow and get my insights on the accuracy of models and limitations of predictions.
References:
Claff, T. et al. Structural basis of lipid-mediated activation of G-protein coupled receptor GPR55. Nat Comm (under revision)
Kung J. E. et al. Disulfide constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM. bioRxiv (2024). https://doi.org/10.1101/2024.05.10.593593
Jumper, J., Evans, R., Pritzel, A. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). https://doi.org/10.1038/s41586-021-03819-2
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871-876 (2021). https://doi.org/10.1126/science.abj8754
The Nobel Prize in Chemistry 2024. NobelPrize.org. Nobel Prize Outreach AB 2024. Fri. 20 Dec 2024. https://www.nobelprize.org/prizes/chemistry/2024/summary/
Edich, M. et al. The impact of AlphaFold2 on experimental structure solution. Faraday Discuss 240, 184-195 (2022). https://doi.org/10.1039/D2FD00072E
Gildea, J. et al. Embedding AI in the Protein Crystallography Workflow. Synchrotron Radiation News 35, 51–54 (2022). https://doi.org/10.1080/08940886.2022.2114723
Goverde, C.A. et al. Computational design of soluble and functional membrane protein analogues. Nature 631, 449–458 (2024). https://doi.org/10.1038/s41586-024-07601-y
Deneke, V.E. et al. A conserved fertilization complex bridges sperm and egg in vertebrates. Cell 187, 7066-7078.e22 (2024). https://doi.org/10.1016/j.cell.2024.09.035
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv (2022). https://doi.org/10.1101/2021.10.04.463034
Gilbert, M.A.G. et al. CryoET of β-amyloid and tau within postmortem Alzheimer’s disease brain. Nature 631, 913–919 (2024). https://doi.org/10.1038/s41586-024-07680-x
Petzold, G. et al. Mining the CRBN Target Space Redefines Rules for Molecular Glue-induced Neosubstrate Recognition. bioRxiv (2024). https://doi.org/10.1101/2024.10.07.616933
Panei, F.P., Gkeka, P. & Bonomi, M. Identifying small-molecules binding sites in RNA conformational ensembles with SHAMAN. Nat Commun 15, 5725 (2024). https://doi.org/10.1038/s41467-024-49638-7