3decision® in the Drug Discovery Pipeline
3decision® completes your Computer-Aided Drug Design tool stack and supports the entire SBDD teams with:
Target assessment: Quickly build the list of targets and evaluate their druggability.
Lead optimization: Easily share and access new structures, also quickly perform structure comparison & overlay
Off-targets evaluation: perform early toxicology assessment by searching for similar ligands or pockets
Working with Biologics: Automatic antibody sequence annotation, detecting CDR regions and loops in contact with antigens, and characteristics of antibodies (heavy/light chain, etc.).
Get a more detailed overview of 3decision® characteristics by looking at the three use cases
Case 1: Early stages of a new project
Search and download existing structures on the target and homologous proteins.
Find druggable binding sites.
Identify potential off-targets.
Find existing binding chemical matter for your target or related proteins (homologous, off-targets).
Download superimposed structures to use them in modeling tools to build pharmacophores and/or run virtual screening campaigns.
No structures for the target yet? Search for homologous proteins and identify the best templates to build homology models from with some visual aid.
Search similar proteins and extract crystallization parameters.
Modelers might run molecular dynamics with their favourite software and upload different model structures that represent the conformational flexibility of the protein.
Developing in-vitro, in-vivo assays? Check mutations in the binding site between species in 3D to avoid undesired surprises.
Case 2: Lead Optimization Loop
Upload a new structure: new holo-structure resolved, new docking model, new homology model.
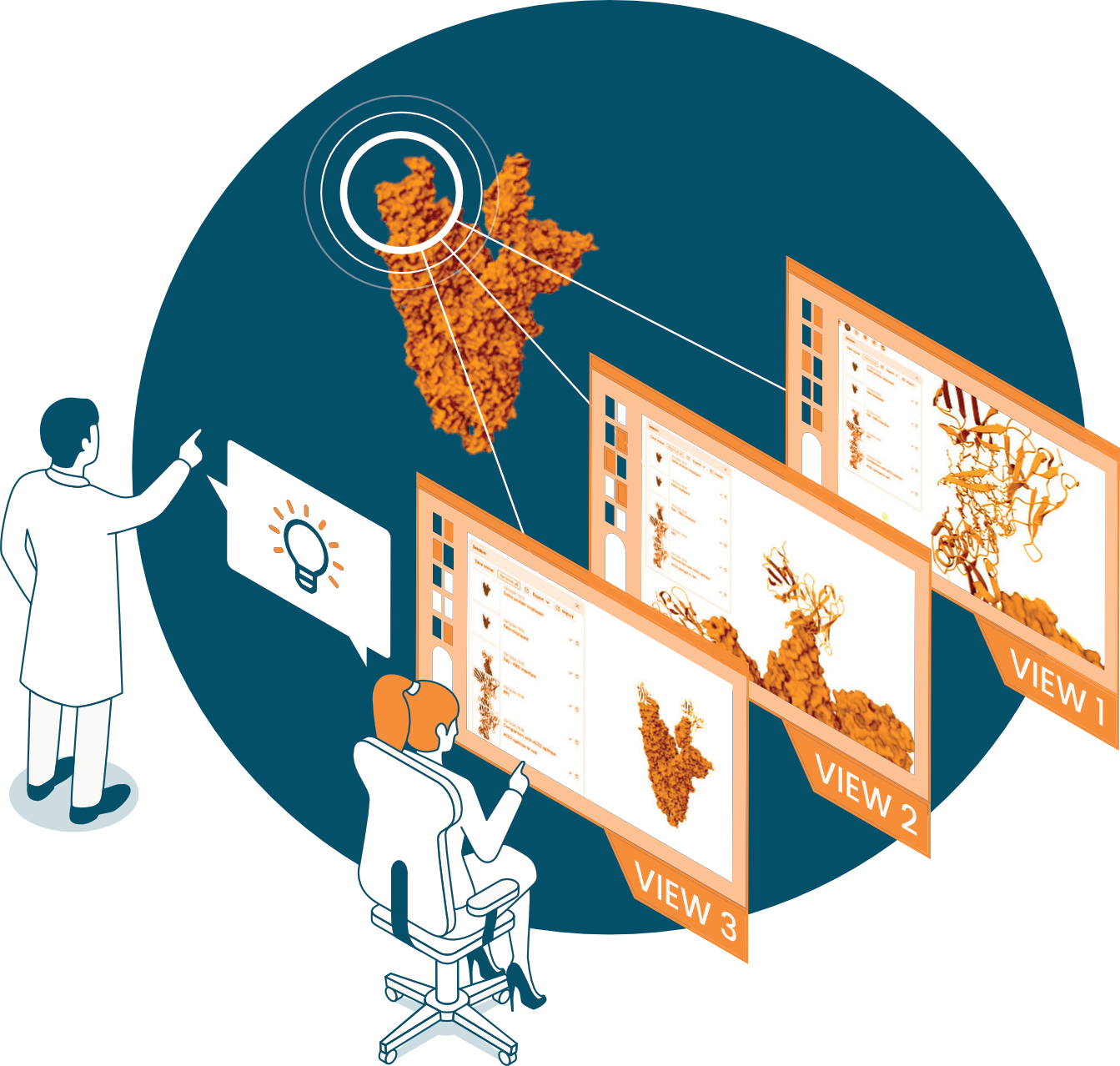
Prepare a 3D scene comparing this new structure with the previous ones in the project to highlight differences of potential interest. For example, a new subpocket opens, giving room to add new functional groups to the ligand.
Compare against off-target structures to reveal mutations in the binding site that could be exploited to gain specificity.
Share the scene with your colleagues using a simple URL link. This will serve as a basis for discussion and decision-making.
At some point, new molecules will be synthesized, and new crystal structures will be resolved, restarting the loop in this way.
Case 3: Collaborate in Drug Discovery Project
Use 3decision® as a common ground for analysis and ideation for drug discovery
Organize structures, models and ligand collections in projects
Find and reuse ideas created in other projects and research programs
Export analyses and data to common data formats
Integrate 3decision into your IT ecosystem via flexible REST APIs