β-catenin
March 2023
β-catenin
The accumulation of β-catenin in cell nucleus is associated with the initiation and progression of various cancer types. This derives from the hyperactivation of the Wingless/integrase-1 (Wnt)/β-catenin pathway. The β-catenin levels are controlled by the action of many other regulatory proteins; therefore β-catenin is engaged in numerous functional protein-protein interactions (PPIs).
Targeting β-catenin PPIs as a therapeutic strategy has been extensively investigated, but it has proven to be very challenging both with small-molecules and biologics. So far, there are no clinically approved drugs for this biological target, and it had been considered “undruggable” for a long time.
However, novel drug modalities have emerged: α-helical stapled peptides (peptides chemically constrained, also called Helicons) have proved effective as inhibitors of PPIs. Using this approach, several new therapeutics targeting β-catenin interactions with other protein partners have been developed.
Very recently, a group from FogPharma developed a new methodology to identify novel binding sites for α-helical peptides. They found and structurally characterized a new Helicon binding site on β-catenin, blocking PPIs with its regulatory protein TCF4, showing their method’s value in discovering novel therapeutic strategies for challenging targets.
The limit of Helicons in drug discovery was that they could only be used to target proteins for which α-helical recognition sites had been previously characterized. So, there was no general method for knowing if a helical peptide could bind to a protein target. To expand the applicability of Helicon technology, the scientists developed a screening method - based on phage display and next-generation sequencing - to identify protein targets for α-helical peptides.
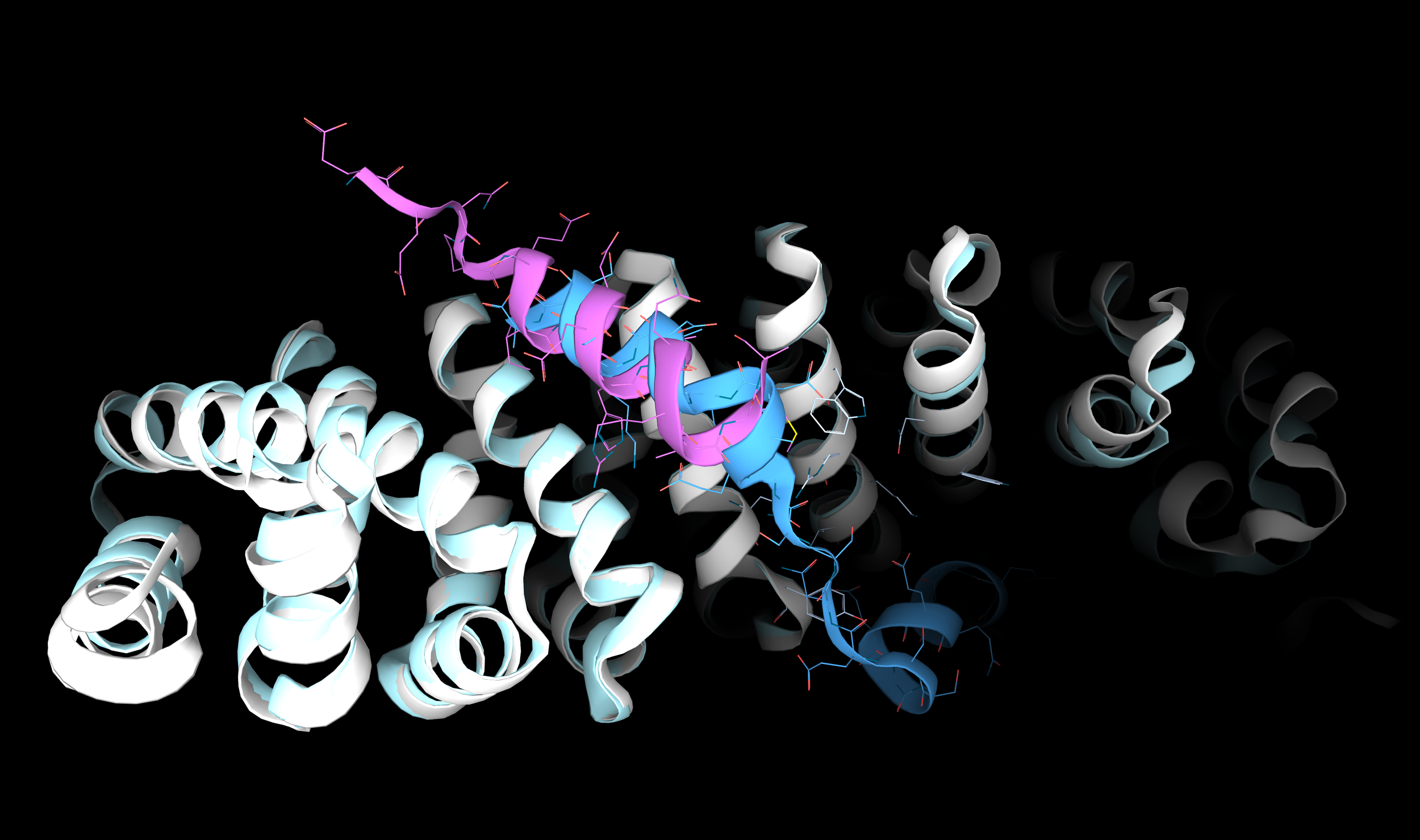
To validate their technique, they used β-catenin because it has a known pocket that binds α-helix from two regulatory proteins: Axin and the TCF4 transcription factor (Image 1).
Image 1. 3D structure of β-catenin interacting with the α-helical portion of two regulatory proteins: Axin (purple; PDB: 1QZ7) and TCF4 (blue; PDB: 1JDH). The picture is produced with the 3decision® software.
Applying their screening method, they identified a set of Helicons peptides that targeted the expected binding site, since they competed with both Axin and TCF4 - the natural binders. From the structural analysis of this cluster of peptides, they not only validated the bio-mimetic binding mode of Helicons, but they identified another binding modality. In fact, some of the binder-peptides (e.g., Helicon FP01567) interacted with the β-catenin surface with similar side chain contacts as the natural binder Axin, but with an inverted N-to-C orientation of the α-helix (Image 2).
Image 2. Protein-protein interactions network between β-catenin and: A) Helicon FP01567 (pink; PDB: 7uwi) and B) Axin (purple; PDB: 1QZ7). The protein-protein interactions are calculated with the 3decision® software.
Unexpectedly, the screening also revealed a further pool of peptides that had yet another binding behavior: the peptides only inhibited TCF4 binding but not Axin. To elucidate the mechanism of inhibition, scientists produced the x-ray structure of β-catenin in complex with Helicon FP05874. They observed that, in this case, the peptide bounded to the protein surface at a completely different location, very distant from the Axin binding site, but at a position that could interfere with TCF4 (Image 3). Targeting this newly discovered binding site allows for selectivity by inhibiting TCF4 interactions but not Axin.
Image 3. Superposition of 3D structures of β-catenin in complex with Helicon FP05874 (yellow; PDB:7uwo), Axin (purple; PDB: 1QZ7) and TCF4 (blue; PDB: 1JDH) to locate the difference in the binding site. For simplicity, the chain of the β-catenin is not displayed for Axin and TCF4. The figure is produced with the 3decision® software.
The combination of this new methodology and structural insights elucidated both a novel binding modality and an unprecedently observed binding site, that could be exploited for the development of novel drugs. This approach has great potential to drive the discovery of innovative therapeutic strategies for challenging protein targets.
BONUS
With 3decision you can calculate protein-protein interactions on-the-fly between two different proteins or between a protein and a peptide. In this way, in just a few clicks, you will be able to explore the network of interactions and spot the critical residues involved.
Reference:
Li K, Tokareva OS, Thomson TM, Wahl SCT, Travaline TL, Ramirez JD, Choudary SK, Agarwal S, Walkup WG 4th, Olsen TJ, Brennan MJ, Verdine GL, McGee JH. De novo mapping of α-helix recognition sites on protein surfaces using unbiased libraries. Proc Natl Acad Sci U S A. 2022 Dec 27;119(52):e2210435119. doi: 10.1073/pnas.2210435119. Epub 2022 Dec 19. PMID: 36534810; PMCID: PMC9907135.