Serotonin receptors (5-HT receptors)
November 2023
Serotonin receptors (5-HT receptors)
Serotonin receptors are a family of proteins that bind to serotonin (also known as 5-hydroxytryptamine, 5-HT), a neurotransmitter that modulates various physiological functions, including mood regulation, appetite, sleep, and cognition. Because of this, drugs targeting serotonin receptors are attractive for the treatment of a wide range of diseases, such as schizophrenia, depression, migraine, and obesity.
This family counts fourteen different protein receptor subtypes, all of which are class A G-protein coupled receptors except for one (5H-T3). Since most of them are structurally very similar, many of the existing drugs usually interact with multiple serotonin receptors, causing adverse effects. This poses the challenge of designing receptor-selective drugs.
Recently, the 3D protein structures for all the serotonin receptor GPCR subtypes were determined, enabling to reveal how protein-ligand recognition occurs at the molecular level for each family member.
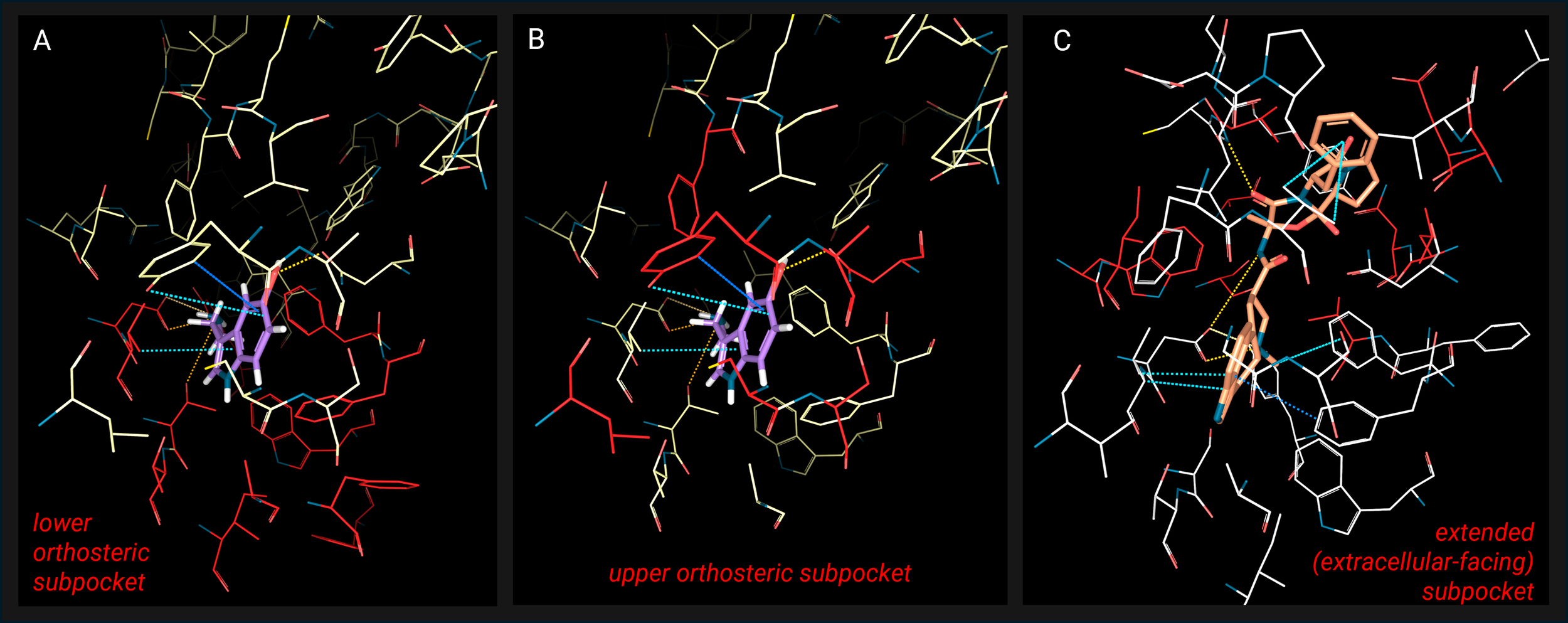
To determine the selectivity hotspots, scientists exploited the available 40 structures of ligand-serotonin receptor complexes. From structural and sequence analysis of these structures, they identified a set of 29 binding site residue positions that are crucial for binding, which define 3 ligand-binding subpockets (Image 1):
“lower orthosteric pocket” binding serotonin’s indole and amine (Image 1A)
“upper orthosteric pocket” interacting with serotonin’s 5-OH group and further stabilizing the indole (Image 1B)
“extended pocket”, extracellular-facing andengaging with larger ligands (Image 1C)
The nature of the protein-ligand interactions is prevalently non-specific, with van der Waals contacts making up 42% of all the contacts, and only 8% of the interactions being polar. This means the ligand recognition is largely due to shape-fit in the binding pocket, and only in part by directed and specific molecular interactions.
Image 1. 3D structure of the binding site of serotonin receptors, with representation of three distinct ligand-binding subpockets: A) 5-HT4 in complex with serotonin (PDB: 7XTA; protein in yellow, ligand in purple), in red are highlighted the residues composing the lower orthosteric subpocket; B) same structure as A, in red are the residues composing the upper orthosteric subpocket; C) 5-HT2C in complex with ergotamine (PDB: 6BQG; protein in white, ligand in orange), in red the extended (extracellular-facing) subpocket. Images created and receptor-ligand contacts calculated with the 3decision® software.
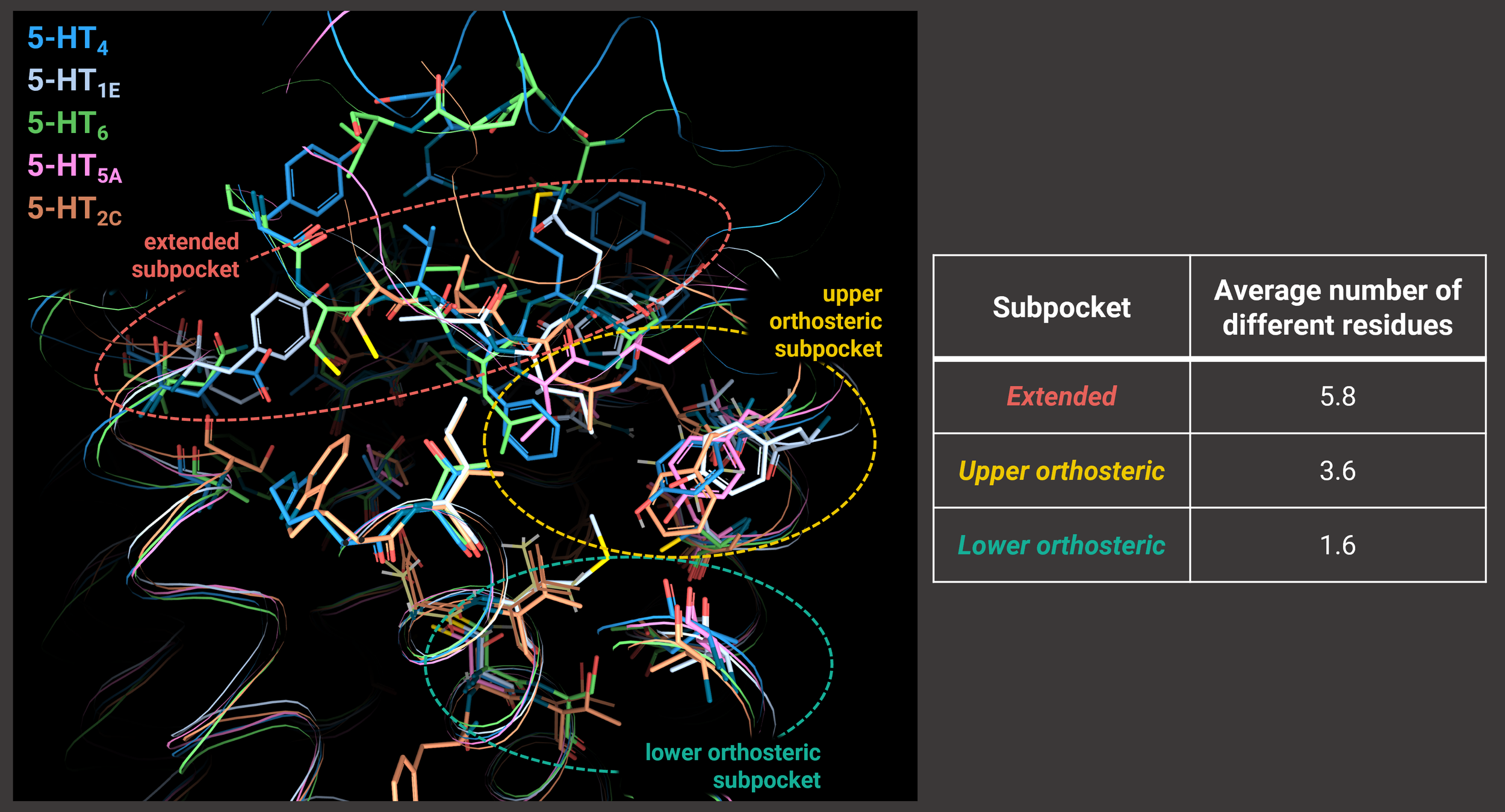
The scientist worked on identifying trends in the similarity of the ligand-binding pockets across the serotonin receptor family, focusing on the previously identified 29 ligand-binding receptor positions. They highlighted that 19 out of 29 of these positions differ in the nature of the amino acid, with the higher diversity found in the lower orthosteric (1.6), followed by the upper orthosteric (3.6), and then extended (5.8) subpocket (Image 2). In this way, they defined a map of these 19 “selectivity hotspots”, that govern the selectivity in the serotonin receptor family.
Image 2. Overlap of the ligand-binding pocket for complexes of the receptors: 5-HT4 (PDB: 7xta, blue), 5-HT1E (PDB: 7e33, white), 5-HT6 (PDB: 7ys6, green), 5-HT5A (PDB: 7um5, pink) and 5-HT2C (PDB: 8dpf, orange). Ligands are hidden for a clearer view. The picture is produced using the 3decision® highlight mode, which only shows the different residues of the superposed proteins. It can be easily seen that the number of residues shown is higher in the lower orthosteric subpocket (in the green circle) due to the higher diversity with respect to the extended subpocket (in the red circle).
For instance, the lower orthosteric pocket shows up to 6 selectivity hotspot residues, that define a shallow and polar subpocket in 5-HT1A-B, 5-HT1D-F, 5-HT5A, and 5-HT7 subtypes, but deeper and hydrophobic in 5-HT2A-C, 5-HT4, and 5-HT6. Ligands carrying hydrophilic groups can be better allocated in this region by the subtypes showing a subpocket with more hydrophilic character. For instance, the ligand sumatriptan shows an average 130-fold preference for the family members with a less hydrophobic character (5-HT1A-B, 5-HT1D-F, 5-HT5A, and 5-HT7) over the other receptors.
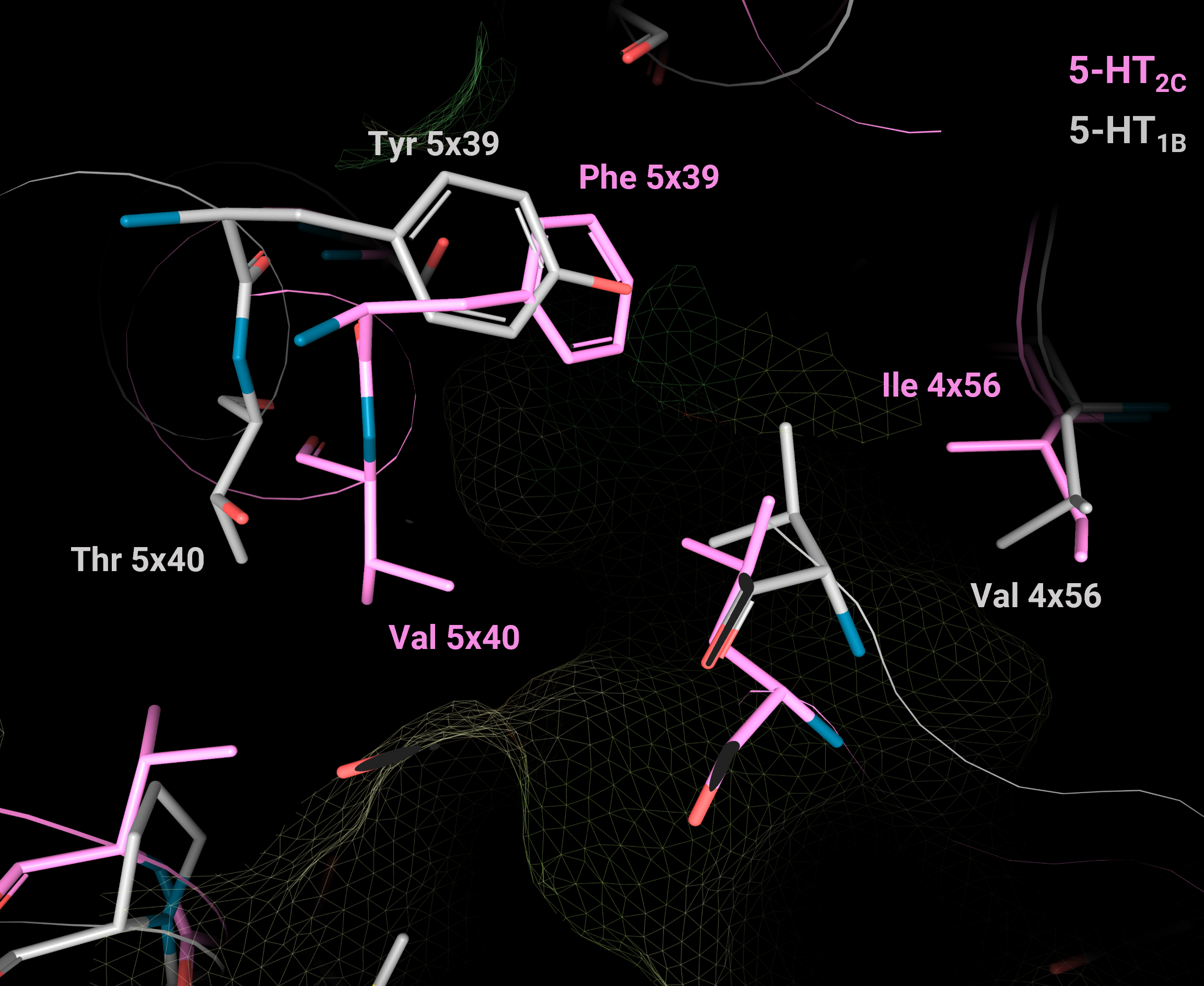
Image 3. Selectivity hotspots in the upper orthosteric pocket between receptors 5-HT2C (PDB: 6BQG, in pink) and 5-HT1B (PDB: 7C61, in grey). 5-HT2C shows a higher hydrophobic character of this pocket (Ile 4x56, Phe 5x39, Val 5x40) compared to the residues at the corresponding position in 5-HT1B (Val 4x56, Tyr 5x39, Thr 5x40). The picture is produced using the 3decision® highlight mode.
Even if the extracellular-facing extended subpocket is the least conserved, it has been less targeted by the thus-far-developed ligands, which only interact with a few residues in this region. The selectivity hotspots in this region are - at the moment - largely unexploited.
This selectivity map and the structural considerations reported in this review could aid the design of novel drugs with increased selectivity and exploring unexploited hotspot regions. This work shows the pivotal contribution of 3D structures in finding new strategies for developing safer therapeutics.
[BONUS: Pocket Similarity Search]
6bqg-Matching ratio: 0.2:V135 T139 A222 F328 S138 G218 S219 F214 F327 D134 L209 V215 N331 V185
SELECTIVITY BASIS & CONCLUSION
Of the total 19 residue positions constituting ligand selectivity hotspots, 13 have experimental evidence whereas six seem unexploited. To aid design of selective ligands, the selectivity map presents receptor-specific sequence signatures along with suggested favorable ligand features. Furthermore, our analysis of selectivity mechanisms (previous section) revealed that selectivity is often achieved via unfavorable contacts in the off-targets – suggesting that extending ligands into sites where the target has unique small residues is a highly efficient strategy.
The size and flexibility of this pocket can allow ligands to find other interactions where the target residue is unique and limits the second strategy of unfavorable off-target contacts. Hence, we need to learn how to better utilize the selectivity hotspots in the extended pocket. Studies are needed to determine the efficiency of, for example, different interactions/bonds and conformationally restriction and increased size of ligands.
POCKETs
Selectivity hotspots in the lower orthosteric pocket [3x32, x33, x36, x37, x40 | 5x461, x47, x44, x48, x51, x52 | 7x42]
Diverse 3×33 and 3×36 residues contribute selectivity for the 5-HT2A–C subfamily
3x33 example
Lorcaserin > target 2C, selectivity over 1A, 3, 4C, 5A, 6, 7. 3x33:Valine is crucial for selectivity. Superposition of Lorcaserin:5HT2C complex (PDB: 8DPF-pocket 1) with 1F (Ile – PDB: 7EXD) and 1E (Met - 7E33) steric clashes between ligand and receptor > 3x33 is a selectivity hotspot
Selectivity hotspots in the upper orthosteric pocket [4x56, 45x52 | 5x39, x40, x43, x44 | 6x55]
The upper orthosteric pocket is less conserved than the lower orthosteric pocket > 6 selectivity hotspots > selective ligand binding
Larger subpocket formed by 4x56, 5x39, 5x40, 5x43:
1A, 1B, 1D, 1E, 1F, 5A, 7: shallow and polar
2A-C, 4, 6: deeper and hydrophobic
Example: Sumatriptan 130-fold highe affinity to subfamily 1 over 2
EXTENDED POCKET [2x60, x63, | 3x28, x29 | 45x51 | 6x58 | 7x31, 34, 35, 38]
The extended pocket is the most diverse and offers ten selectivity hotspots. This pocket extends to the extracellular interface (Figure 3A) and can accommodate large ligand substituents on the amine nitrogen, the extended pocket has the fewest ligand interactions (Figure 3B) and mutations. this pocket has an unexploited potential to contribute selectivity to new ligands with extended scaffolds.
ALLOSTERIC
Advances in the structure-based design of allosteric ligands [47–49] open for binding solely outside of the orthosteric pocket of serotonin.
SELECTIVITY BASIS
Selectivity can arise through favorable on-target interactions or unfavorable off-target contacts, such as steric obstruction and electrostatic repulsion.
Our analysis of selectivity mechanisms (previous section) revealed that selectivity is often achieved via unfavorable contacts in the off-targets – suggesting that extending ligands into sites where the target has unique small residues is a highly efficient strategy.
CONCLUSION
This provides a foundation to generate structure-selectivity relationships for a larger number of serotonergic ligands, as well as other GPCRs, and hypotheses for future medicinal chemistry and mutagenesis studies – ultimately leading to more selective tool compounds and safer drugs.
This study has presented a detailed mapping of ligand selectivity-inducing hotspots among 29 binding site residue positions across the 12 human serotonin receptors (Figure 4). This integrative map was generated from structural and sequence information, enriched with experimental and predicted ligand binding modes, and used to rationalize observed ligand-binding affinities and mutagenesis effects. Of the total 19 residue positions constituting ligand selectivity hotspots, 13 have experimental evidence, whereas six seem unexploited.
Selectivity and pocket similarity
Grouping receptors based on their similarity in the ligand-binding pocket rather than overall sequence can pinpoint the most challenging targets for the design of selective ligands
Focusing on the 29 ligand-binding residue positions. The serotonin receptor family is divided into three groups of highly similar receptors:
(i) 1A, 5A and 7,
(ii) 1B, D, E, F
(iii) 2A, 2B, 2C
(iv) two outliers 4 and 6.
Observed off-target activities:
pairs 5-HT1B/5-HT1D
5-HT2A/5-HT2C
For which development of subtype-selective ligands has proven particularly challenging
References for off-target:
The potential selectivity hotspots are largely unexploited by structure ligands.
| PDB | Serotonin receptor | Active/Inactive state | Ligand | 3dec issue |
|---|---|---|---|---|
| 7E2Y | 5-HT1A | Active | Serotonin | |
| 6G79 | 5-HT1B | Active | Donitriptan | |
| 5V54 | 5-HT1B | Inactive | Methiothepin | |
| 7E32 | 5-HT1D | Active | Serotonin | |
| 7E33 | 5-HT1E | Active | BRL54443 | |
| 7EXD | 5-HT1F | Active | Lasmiditan | |
| 6WHA | 5-HT2A | Active | 25-CN-NBOH | |
| 6A94 | 5-HT2A | Inactive | Zotepine | |
| 7SRR | 5-HT2B | Active | LSD | |
| 6DRX | 5-HT2B | Inactive | Lisuride | |
| 8DPF | 5-HT2C | Active | Lorcaserin | |
| 6BQH | 5-HT2C | Inactive | Ritansein | |
| 7XT8 | 5-HT4 | Active | Serotonin | No chain mapping |
| 7UM5 | 5-HT5A | Active | 5-CT | |
| 7UM4 | 5-HT5A | Inactive | AS2674723 | |
| 8JLZ | 5-HT6 | Active | ST1936 | No chain mapping |
| 7XTC | 5-HT7 | Active | 5-CT | No chain mapping |
| Method | PDB ID | Res. (Å) | Pref. chain | State | Ligand | Ligand modality | Sign. prot. | Fusion protein | Anti-body | Ref | Year | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5-HT1A | cryo-EM | 7E2X | 3.0 | R | Active | - (apo) | - | Gi1 | - | - | [6] | 2021 |
| cryo-EM | 7E2Z | 3.1 | R | Active | Aripiprazole | Agonist (partial) | Gi1 | - | - | [6] | 2021 | |
| cryo-EM | 7E2Y | 3.0 | R | Active | Serotonin | Agonist | Gi1 | - | - | [6] | 2021 | |
| 5-HT1B | X-ray | 7C61 | 3.0 | A | Inactive | Ergotamine | Antagonist | - | BRIL | IgG heavy chain | [8] | 2020 |
| cryo-EM | 6G79 | 3.8 | S | Active | Donitriptan | Agonist | Go | - | - | [10] | 2018 | |
| X-ray | 5V54 | 3.9 | A | Inactive | Methiothepin | Antagonist | - | BRIL | - | [17] | 2018 | |
| X-ray | 4IAR | 2.7 | A | Inactive | Ergotamine | Agonist | - | BRIL | - | [18] | 2013 | |
| X-ray | 4IAQ | 2.8 | A | Inactive | Dihydroergotamine | Agonist | - | BRIL | - | [18] | 2013 | |
| 5-HT1D | cryo-EM | 7E32 | 2.9 | R | Active | Serotonin | Agonist | Gi1 | - | scFv16 | [6] | 2021 |
| 5-HT1E | cryo-EM | 7E33 | 2.9 | R | Active | BRL-54443 | Agonist | Gi1 | - | scFv16 | [6] | 2021 |
| 5-HT1F | cryo-EM | 7EXD | 3.4 | R | Active | Lasmiditan | Agonist | Gi1 | BRIL | scFv16 | [5] | 2021 |
| 5-HT2A | X-ray | 7RAN | 3.5 | A | Active | R-69 | Agonist | Gq | - | - | [19] | 2022 |
| X-ray | 7WC7 | 2.6 | A | Inactive | Lisuride | Agonist | - | BRIL | - | [20] | 2022 | |
| X-ray | 7WC4 | 3.2 | A | Inactive | Serotonin | Agonist | - | BRIL | - | [20] | 2022 | |
| X-ray | 7WC8 | 2.5 | A | Inactive | Lumateperone | Agonist | - | BRIL | - | [20] | 2022 | |
| X-ray | 7WC6 | 2.6 | A | Inactive | LSD | Agonist | - | BRIL | - | [20] | 2022 | |
| X-ray | 7WC5 | 3.2 | A | Inactive | Psilocin | Agonist | - | BRIL | - | [20] | 2022 | |
| X-ray | 7WC9 | 2.5 | A | Inactive | IHCH-7086 | Agonist | - | BRIL | - | [20] | 2022 | |
| X-ray | 7VOE | 2.9 | A | Inactive | Aripiprazole | Agonist (partial) | - | BRIL | - | [21] | 2021 | |
| X-ray | 7VOD | 3.3 | A | Inactive | Cariprazine | Agonist (partial) | - | BRIL | - | [21] | 2021 | |
| X-ray | 6WH4 | 3.4 | C | Inactive | Methiothepin | Inverse agonist | - | BRIL | - | [7] | 2020 | |
| X-ray | 6WGT | 3.4 | B | Inactive | LSD | Agonist | - | BRIL | - | [7] | 2020 | |
| cryo-EM | 6WHA | 3.4 | A | Active | 25CN-NBOH | Agonist | Gq | BRIL | scFv16 | [7] | 2020 | |
| X-ray | 6A94 | 2.9 | A | Inactive | Zotepine | Antagonist | - | BRIL | - | [22] | 2019 | |
| X-ray | 6A93 | 3.0 | A | Inactive | Risperidone | Antagonist | - | BRIL | - | [22] | 2019 | |
| 5-HT2B | X-ray | 6DRZ | 3.1 | A | Intermediate | Methysergide | Antagonist | - | BRIL | - | [23] | 2018 |
| X-ray | 6DRY | 2.9 | A | Intermediate | Methylergonovine | Agonist | - | BRIL | - | [23] | 2018 | |
| X-ray | 6DRX | 3.1 | A | Intermediate | Lisuride | Antagonist | - | BRIL | - | [23] | 2018 | |
| X-ray | 6DS0 | 3.2 | A | Intermediate | LY266097 | Antagonist | - | BRIL | - | [23] | 2018 | |
| X-ray | 5TUD | 3.0 | D | Active | Ergotamine | Agonist | - | BRIL | P2C2–Fab | [12] | 2017 | |
| X-ray | 5TVN | 2.9 | A | Intermediate | LSD | Agonist | - | BRIL | - | [13] | 2017 | |
| X-ray | 4NC3 | 2.8 | A | Intermediate | Ergotamine | Agonist | - | BRIL | - | [24] | 2013 | |
| X-ray | 4IB4 | 2.7 | A | Intermediate | Ergotamine | Agonist | - | BRIL | - | [25] | 2013 | |
| cryo-EM | 7SRQ | 2.7 | R | Intermediate | LSD | Agonist | - | - | - | [26] | 2022 | |
| cryo-EM | 7SRR | 2.9 | R | Active | LSD | Agonist | Gq | - | - | [26] | 2022 | |
| cryo-EM | 7SRS | 3.3 | R | Active | LSD | Agonist | Barr1 | - | - | [26] | 2022 | |
| 5-HT2C | X-ray | 6BQH | 2.7 | A | Inactive | Ritanserin | Inverse agonist | - | BRIL | - | [1] | 2018 |
| X-ray | 6BQG | 3.0 | A | Active | Ergotamine | Agonist | - | BRIL | - | [1] | 2018 | |
| cryo-EM | 8DPF | 2.8 | A | Active | Lorcaserin | Agonist | Gq | - | - | [4] | 2022 | |
| cryo-EM | 8DPG | 3.6 | A | Active | Psilocin | Agonist | - | - | - | [4] | 2022 | |
| cryo-EM | 8DPH | 3.2 | A | Active | Lorcaserin | Agonist | - | - | - | [4] | 2022 | |
| cryo-EM | 8DPI | 3.4 | A | Active | Lorcaserin | Agonist | - | - | - | [4] | 2022 | |
| 5-HT4 | cryo-EM | 7XT8 | 3.1 | R | Active | Serotonin | Agonist | Gs | BRIL | Nb35 | [9] | 2022 |
| cryo-EM | 7XT9 | 3.2 | R | Active | Serotonin | Agonist | Gs | - | - | [9] | 2022 | |
| cryo-EM | 7XTA | 3.2 | R | Active | Serotonin | Agonist | Gi1 | - | - | [9] | 2022 | |
| 5-HT5A | X-ray | 7UM4 | 2.8 | A | Inactive | AS2674723 | Antagonist | - | PGS | - | [27] | 2022 |
| cryo-EM | 7UM5 | 2.7 | A | Active | 5-CT | Agonist | Go | - | scFv16 | [27] | 2022 | |
| cryo-EM | 7UM6 | 2.8 | A | Active | Lisuride | Agonist (partial) | Go | - | - | [27] | 2022 | |
| cryo-EM | 7UM7 | 2.8 | A | Active | Methylergometrine | Agonist | Go | - | - | [27] | 2022 | |
| cryo-EM | 7X5H | 3.1 | R | Active | 5-CT | Agonist | Gi1 | - | - | [28] | 2022 | |
| 5-HT6 | cryo-EM | 7XTB | 3.3 | R | Active | Serotonin | Agonist | Gs | BRIL | Nb35 | [9] | 2022 |
| cryo-EM | 7YS6 | 3.0 | A | Active | Serotonin | Agonist | Gs | - | scFv16 | -- | 2023 | |
| cryo-EM | 8JLZ | 3.1 | R | Active | ST-1936 | Agonist | Gs | - | Nb35 | [29] | 2023 | |
| 5-HT7 | cryo-EM | 7XTC | 3.2 | R | Active | 5-CT | Agonist | Gs | BRIL | Nb35 | [9] | 2022 |