SARS-CoV-2 S protein
March 2021
Since the beginning of this month, the milestone of more than 1000 SARS-CoV-2 publicly available protein structures has been reached. It confirms a remarkable effort in structural biology to increase the understanding of the infection mechanism and support the search for treatment against COVID-19.
The S (spike) protein, one of the SARS-CoV-2 structural proteins, has now over 300 resolved structures registered in the RCSB PDB. This number reflects the huge interest in this particular protein for COVID-19 vaccine development. Since the appearance of the first SARS-CoV-2 “variant” the importance of correctly mapping the amino acid sequence to the protein 3D structure has been highlighted.
For example, in the so-called “English variant”, the asparagine residue has been replaced with tyrosine on the position of 501 in the Spike protein’s receptor-binding domain (RBD). This part of the protein is exposed on the outside of the virus, and the N501Y mutation can therefore affect the binding of antibodies. New and more virulent SARS-Cov2 variants raise questions about the efficiency of current vaccines.
In the 3decision structural knowledge base, you can instantly access all the publicly available SARS-CoV-2 protein structures. You can also upload and store proprietary structures and homology models. The advanced search options serve to refine the query, filter S protein structures, add to the project, and analyze the sequence-to-structure mapping for structural study.
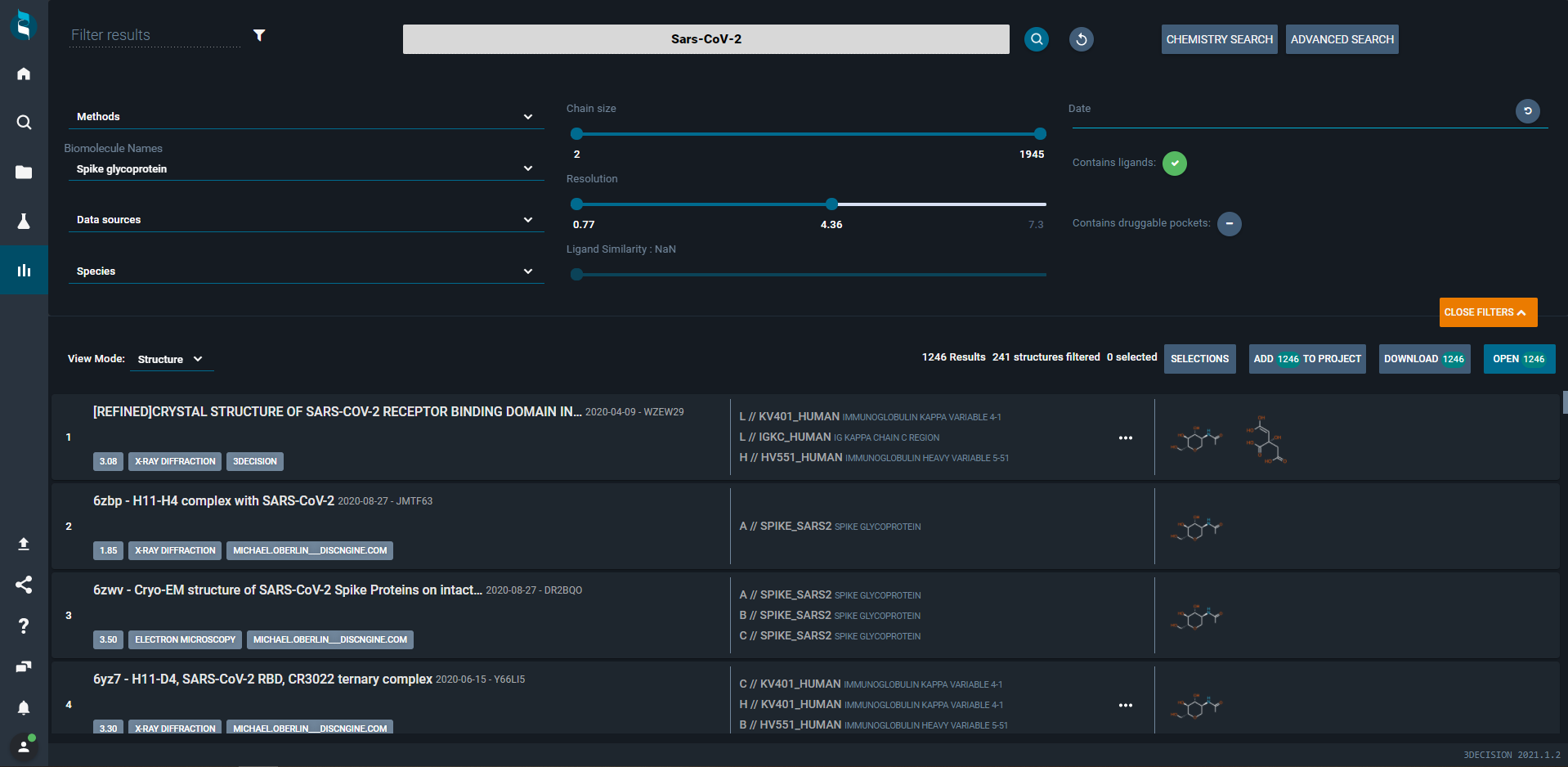
The advanced search feature in 3decision showing filtered structures of Spike protein of SARS-CoV-2
Check out some of the articles on Sars-Cov-2 structural study from our team :
2019-ncov-structurally-speaking
SARS-CoV-2 - From the viral genome to protein structures
