Getting ready for AlphaFold with 3decision®
The most recent development in the structural biology field comes from DeepMind company’s system AlphaFold. Using artificial intelligence, the system can predict the unique three-dimensional shape of a protein with high accuracy - being remarkably close to laboratory experimental results. It is happening for the first time that computational models are considered compatible with experimental methods. Together with the incredible speed of this technique, we could now call such an achievement “The AI breakthrough in protein folding.”
The AlphaFold deep learning approach, once validated, could have enormous potential in the field of structure-based drug discovery and a crucial impact on accelerating the development of novel drugs. Scientists will be able to obtain thousands of yet unknown protein structures of the human body and other organisms quickly and reliably. This would most likely be an additional large source of data for scientific research. On top of modeling, there are parallel improvements in experimental techniques, such as high throughput crystallography screening. New demands on how to properly organize and make the most of structural knowledge will arise, particularly during the earliest stages of drug discovery. It might be very challenging for scientists to maintain the overall track of the project data. Failing to do so and thus being lost in the crowd might lead to missed opportunities and unexpected slips.
Foreseen data avalanche is what, already 5 years ago, inspired scientists at Discngine to start the development of the protein-structure repository - 3decision®. The software can store all relevant structural data from various sources in one place. It integrates structures from the PDB database together with calculated biological assemblies and computational models from the public domain. Moreover, all proprietary in-house structures and models can be added, curated, and analyzed. Protein sequences from UniProt are regularly synchronized in the system too and finally, all additional data linked to the structure or the project can be stored. With such a knowledge base, 3decision® can prevent the possible loss of any valuable project information and minimize human errors. Scientists are able to exploit the full potential of their structural knowledge in an automated and timely manner. The centralized platform is also equipped with advanced structural analytics capabilities to test hypotheses and share the results within the entire drug design team.
The mentioned strengths of 3decision® can prepare scientists for what might come with modeling approaches such as AlphaFold if they reach routine use in the drug industry. There are still many questions to answer, as DeepMind reports. However, we are excited about all the new possibilities that will enable scientists to gain a better understanding of protein structures, especially ones currently difficult to crystallize.
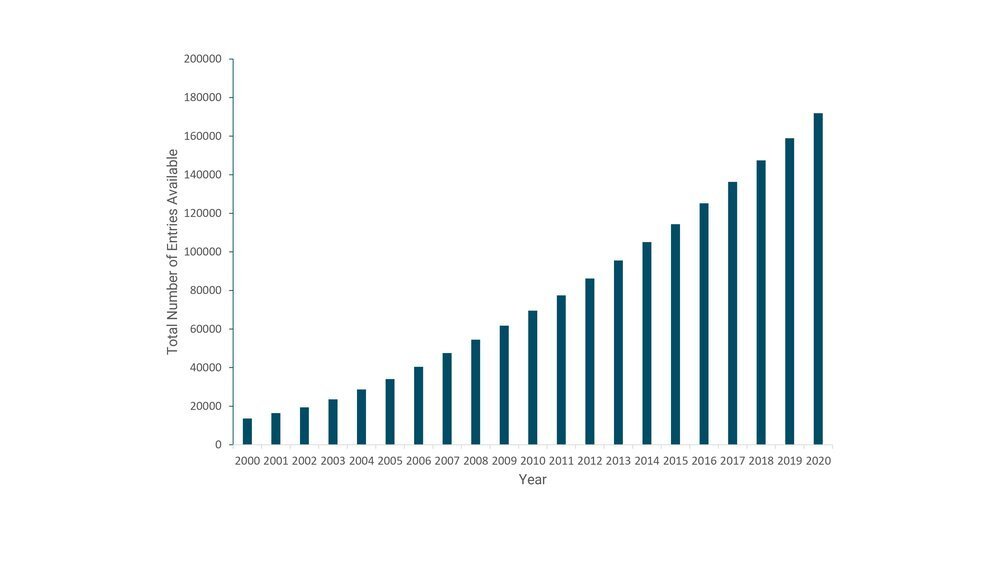
Current number of entries in RCSB PDB
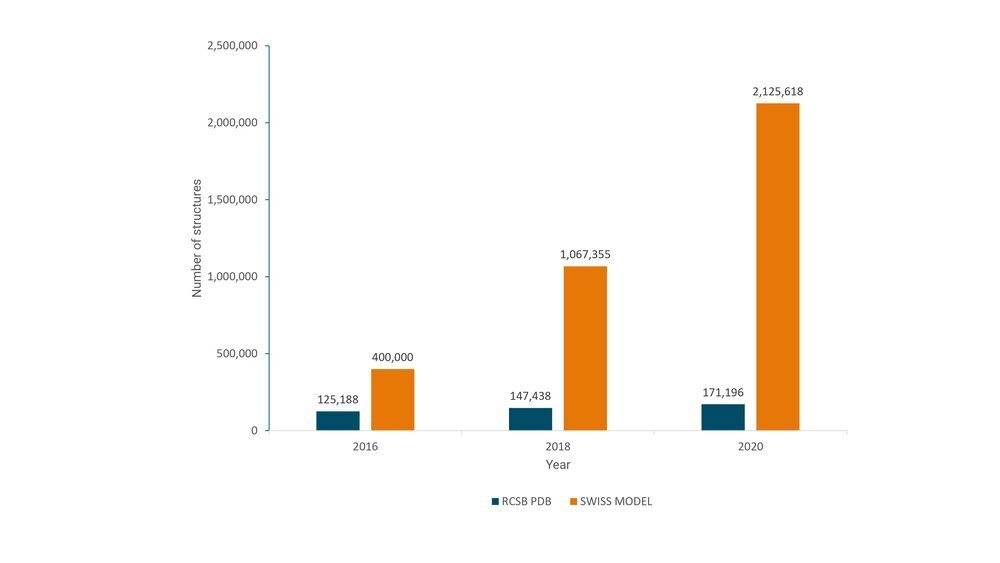
Structural data growth in the last 4 years: Computationally generated models (SWISS MODEL repository) compared with experimentally determined structures in PDB
References:
1- Stefan Bienert, Andrew Waterhouse, Tjaart A. P. de Beer, Gerardo Tauriello, Gabriel Studer, Lorenza Bordoli, Torsten Schwede, The SWISS-MODEL Repository—new features and functionality, Nucleic Acids Research, Volume 45, Issue D1, January 2017, Pages D313–D319, https://doi.org/10.1093/nar/gkw1132
2 - Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modeling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296-W303. doi:10.1093/nar/gky427
3 - https://swissmodel.expasy.org/repository
4 - https://www.rcsb.org/stats/growth/growth-released-structures
5 - https://www.uniprot.org/
6 - https://www.nature.com/articles/d41586-020-03348-4