SARS-CoV-2 - part 2 - From the viral genome to protein structures
After our first announcement yesterday here, let’s try to get an overview of the viral genome. That’s already rather complicated given the current flood of information. So what I gathered here for now is by no means complete, but it gives us a decent starting point.
The bread and butter of every structure-based drug design or drug ID effort is the need for 3D structural information of a target of interest related to the phenotypic outcome we want to alter. In the case of SARS-CoV-2, several experimental structures are already available in the RCSB PDB. In addition, X-ray structures from a large scale fragment screen from the Diamond Light Source are also slowly becoming available on public resources like the RCSB PDB (the entire list is already accessible in 3decision’s main protease project). Popular homology modeling services have also already started to provide several high-quality models for some of the yet not resolved structures of the viral proteome.
But let’s first have a look at what the viral genome actually contains and what the currently pursued therapeutic efforts are.
The genome of SARS-CoV-2
The current status of sequencing data and analysis is available on ExPASy (viralzone) and summarized here below.
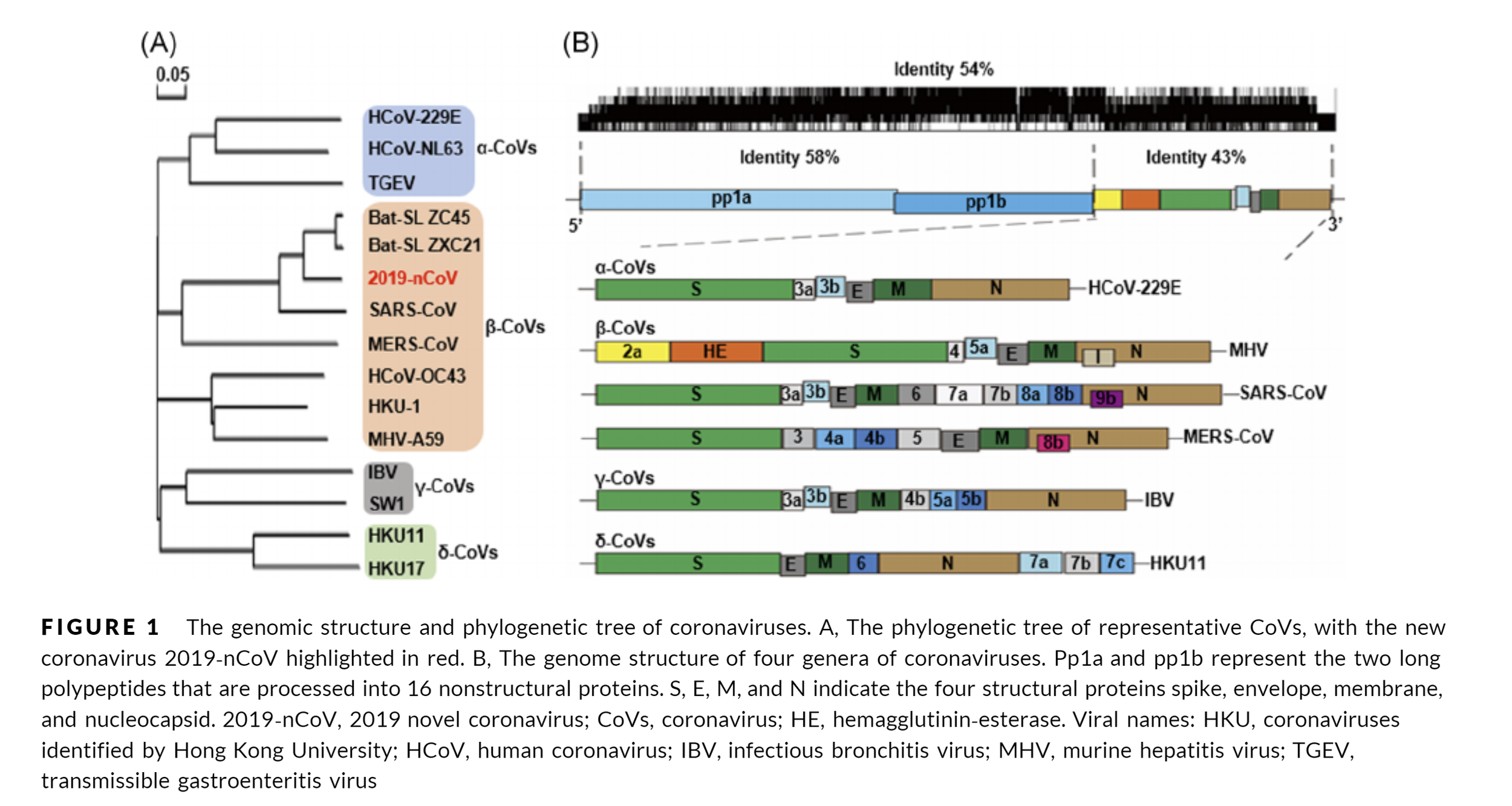
The SARS-CoV-2 genome structure as available on viralzone.expasy.org
Image extracted from a review on coronavirus genome structures from early 2020.
The genome of SARS-CoV-2 is split into two main regions. The first part of the genomic RNA is used as a template to directly translate polyprotein 1a/1ab (pp1a/pp1ab or ORF1a ORF1b), which encodes non-structural proteins (nsps) to form the replication‐transcription complex (RTC).
There is a −1 frameshift between ORF1a and ORF1b, leading to the production of two polypeptides: pp1a and pp1ab. These polypeptides are processed into 16 nsps by the virally encoded main protease (Mpro) - a chymotrypsin‐like protease (3CLpro), that a lot of people are after right now in structure-based drug design efforts), and one or two papain‐like proteases (PLpro). [https://onlinelibrary.wiley.com/doi/epdf/10.1002/jmv.25681].
The second part of the genome encodes four main structural proteins: the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins. Besides these four main structural proteins, different CoVs encode special structural and accessory proteins, such as the HE protein, 3a/b protein, and 4a/b protein (Figure 1B, lower panel). All the structural and accessory proteins are translated from the sgRNAs of CoVs [https://onlinelibrary.wiley.com/doi/epdf/10.1002/jmv.25681].
Among these structural proteins, some are exposed to the outside of the virus:
The lifecycle of the virus
To identify potential targets it is very important to understand the function of its constituents and how the virus integrates a cell, replicates and disseminates afterwards again.
Taken from a review on the previous MERS-CoV outbreak. Apart from a few details, the current SARS-CoV-2 acts rather similarly.
The infection mechanism of MERS-CoV is very well explained in this review. The main difference for now with SARS-CoV-2 is the targeted main receptor on the host cell. SARS-CoV-2 is suspected to bind to the angiotensin-converting enzyme 2 (ACE-2), as opposed to MERS-CoV which binds to dipeptidyl peptidase 4 (DPP4) . However, a lot still has to be discovered and it is possible that other receptors might play a role in the fixation of the virus on the cell. For example, this paper stipulates that the Spike protein of SARS-CoV-2 might actually also expose features known to bind to integrins (the RGD motif). A lot will become available during the next months and years and we’ll try to update the information available here as much as possible.
The previous review also lists several therapeutic approaches. We’ll cover that a bit later here.
Functional annotations of the non-structural protein (nsp)
Let’s have a closer look at the 16 nsps resulting from the digestion of SARS-CoV2’s polypeptides pp1a and pp1ab. We’ll start by getting an overview. We’ll try to go into more detail for each of them in upcoming posts. Here I’ll also state if there already are known experimental structures known for this protein or close homologs in other viruses.
An extract of the non structural proteins (nsp) annotations on the official Uniprot entry of SARS-CoV2’s polyprotein pp1ab. Visualisation from 3decision.
The following
nsp1 (180 amino acids):
Residue 1-180 in orf1ab polyprotein MESLVPGFNEKTHVQLSLPVLQVRDVLVRGFGDSVEEVLSEARQHLKDGTCGLVEVEKGVLPQLEQPYVFIKRSDARTAPHGHVMVELVAELEGIQYGRSGETLGVLVPHVGEIPVAYRKVLLRKNGNKGAGGHSYGADLKSFDLGDELGTDPYEDFQENWNTKHSSGVTRELMRELNGG
Cellular mRNA degradation, inhibiting IFN signaling [https://onlinelibrary.wiley.com/doi/epdf/10.1002/jmv.25681]
Inhibits host translation by interacting with the 40S ribosomal subunit. The nsp1-40S ribosome complex further induces an endonucleolytic cleavage near the 5'UTR of host mRNAs, targeting them for degradation. Viral mRNAs are not susceptible to nsp1-mediated endonucleolytic RNA cleavage thanks to the presence of a 5'-end leader sequence and are therefore protected from degradation. By suppressing host gene expression, nsp1 facilitates efficient viral gene expression in infected cells and evasion from host immune response. [https://swissmodel.expasy.org/repository/species/2697049]
Host translation inhibitor nsp1.
3D-Structures:
nsp2 (638 amino acids):
Residue 181-818 in orf1ab polyprotein AYTRYVDNNFCGPDGYPLECIKDLLARAGKASCTLSEQLDFIDTKRGVYCCREHEHEIAWYTERSEKSYELQTPFEIKLAKKFDTFNGECPNFVFPLNSIIKTIQPRVEKKKLDGFMGRIRSVYPVASPNECNQMCLSTLMKCDHCGETSWQTGDFVKATCEFCGTENLTKEGATTCGYLPQNAVVKIYCPACHNSEVGPEHSLAEYHNESGLKTILRKGGRTIAFGGCVFSYVGCHNKCAYWVPRASANIGCNHTGVVGEGSEGLNDNLLEILQKEKVNINIVGDFKLNEEIAIILASFSASTSAFVETVKGLDYKAFKQIVESCGNFKVTKGKAKKGAWNIGEQKSILSPLYAFASEAARVVRSIFSRTLETAQNSVRVLQKAAITILDGISQYSLRLIDAMMFTSDLATNNLVVMAYITGGVVQLTSQWLTNIFGTVYEKLKPVLDWLEEKFKEGVEFLRDGWEIVKFISTCACEIVGGQIVTCAKEIKESVQTFFKLVNKFLALCADSIIIGGAKLKALNLGETFVTHSKGLYRKCVKSREETGLLMPLKAPKEIIFLEGETLPTEVLTEEVVLKTGDLQPLEQPTSEAVEAPLVGTPVCINGLMLLEIKDTEKYCALAPNMMVTNNTFTLKGG
May play a role in the modulation of host cell survival signaling pathway by interacting with host PHB and PHB2. Indeed, these two proteins play a role in maintaining the functional integrity of the mitochondria and protecting cells from various stresses. [https://swissmodel.expasy.org/repository/species/2697049, https://jvi.asm.org/content/83/19/10314]
3D-Structures:
No known experimental structure
nsp3 (1945 amino acids):
Residue 819-2763 in orf1ab polyprotein APTKVTFGDDTVIEVQGYKSVNITFELDERIDKVLNEKCSAYTVELGTEVNEFACVVADAVIKTLQPVSELLTPLGIDLDEWSMATYYLFDESGEFKLASHMYCSFYPPDEDEEEGDCEEEEFEPSTQYEYGTEDDYQGKPLEFGATSAALQPEEEQEEDWLDDDSQQTVGQQDGSEDNQTTTIQTIVEVQPQLEMELTPVVQTIEVNSFSGYLKLTDNVYIKNADIVEEAKKVKPTVVVNAANVYLKHGGGVAGALNKATNNAMQVESDDYIATNGPLKVGGSCVLSGHNLAKHCLHVVGPNVNKGEDIQLLKSAYENFNQHEVLLAPLLSAGIFGADPIHSLRVCVDTVRTNVYLAVFDKNLYDKLVSSFLEMKSEKQVEQKIAEIPKEEVKPFITESKPSVEQRKQDDKKIKACVEEVTTTLEETKFLTENLLLYIDINGNLHPDSATLVSDIDITFLKKDAPYIVGDVVQEGVLTAVVIPTKKAGGTTEMLAKALRKVPTDNYITTYPGQGLNGYTVEEAKTVLKKCKSAFYILPSIISNEKQEILGTVSWNLREMLAHAEETRKLMPVCVETKAIVSTIQRKYKGIKIQEGVVDYGARFYFYTSKTTVASLINTLNDLNETLVTMPLGYVTHGLNLEEAARYMRSLKVPATVSVSSPDAVTAYNGYLTSSSKTPEEHFIETISLAGSYKDWSYSGQSTQLGIEFLKRGDKSVYYTSNPTTFHLDGEVITFDNLKTLLSLREVRTIKVFTTVDNINLHTQVVDMSMTYGQQFGPTYLDGADVTKIKPHNSHEGKTFYVLPNDDTLRVEAFEYYHTTDPSFLGRYMSALNHTKKWKYPQVNGLTSIKWADNNCYLATALLTLQQIELKFNPPALQDAYYRARAGEAANFCALILAYCNKTVGELGDVRETMSYLFQHANLDSCKRVLNVVCKTCGQQQTTLKGVEAVMYMGTLSYEQFKKGVQIPCTCGKQATKYLVQQESPFVMMSAPPAQYELKHGTFTCASEYTGNYQCGHYKHITSKETLYCIDGALLTKSSEYKGPITDVFYKENSYTTTIKPVTYKLDGVVCTEIDPKLDNYYKKDNSYFTEQPIDLVPNQPYPNASFDNFKFVCDNIKFADDLNQLTGYKKPASRELKVTFFPDLNGDVVAIDYKHYTPSFKKGAKLLHKPIVWHVNNATNKATYKPNTWCIRCLWSTKPVETSNSFDVLKSEDAQGMDNLACEDLKPVSEEVVENPTIQKDVLECNVKTTEVVGDIILKPANNSLKITEEVGHTDLMAAYVDNSSLTIKKPNELSRVLGLKTLATHGLAAVNSVPWDTIANYAKPFLNKVVSTTTNIVTRCLNRVCTNYMPYFFTLLLQLCTFTRSTNSRIKASMPTTIAKNTVKSVGKFCLEASFNYLKSPNFSKLINIIIWFLLLSVCLGSLIYSTAALGVLMSNLGMPSYCTGYREGYLNSTNVTIATYCTGSIPCSVCLSGLDSLDTYPSLETIQITISSFKWDLTAFGLVAEWFLAYILFTRFFYVLGLAAIMQLFFSYFAVHFISNSWLMWLIINLVQMAPISAMVRMYIFFASFYYVWKSYVHVVDGCNSSTCMMCYKRNRATRVECTTIVNGVRRSFYVYANGGKGFCKLHNWNCVNCDTFCAGSTFISDEVARDLSLQFKRPINPTDQSSYIVDSVTVKNGSIHLYFDKAGQKTYERHSLSHFVNLDNLRANNTKGSLPINVIVFDGKSKCEESSAKSASVYYSQLMCQPILLLDQALVSDVGDSAEVAVKMFDAYVNTFSSTFNVPMEKLKTLVATAEAELAKNVSLDNVLSTFISAARQGFVDSDVETKDVVECLKLSHQSDIEVTGDSCNNYMLTYNKVENMTPRDLGACIDCSARHINAQVAKSHNIALIWNVKDFMSLSEQLRKQIRSAAKKNNLPFKLTCATTRQVVNVVTTKIALKGG
Alternative name: papain-like proteinase
Responsible for the cleavages located at the N-terminus of the replicase polyprotein. In addition, PL-PRO possesses a deubiquitinating/deISGylating activity and processes both 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains from cellular substrates. Participates together with nsp4 in the assembly of virally induced cytoplasmic double-membrane vesicles necessary for viral replication. Antagonises innate immune induction of type I interferon by blocking the phosphorylation, dimerisation and subsequent nuclear translocation of host IRF3. Prevents also host NF-kappa-B signaling.[https://swissmodel.expasy.org/repository/species/2697049]. As indicated on the expasy schema, it’s cleaving the polyprotein between nsp1 & 2, nsp2 and itself and itself and nsp4.
Large, multi-domain transmembrane protein, activities include: [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4369385/ general review on coronaviruses]
Ubl1 and Ac domains, interact with N protein
ADRP activity, promotes cytokine expression
PLPro/Deubiquitinase domain, cleaves viral polyprotein and blocks host innate immune response
Ubl2, NAB, G2M, SUD, Y domains, unknown functions
3D-Structures
One of the domains covered by experimental structures
nsp4 (500 amino acids):
Residue 2764-3263 in orf1ab polyprotein KIVNNWLKQLIKVTLVFLFVAAIFYLITPVHVMSKHTDFSSEIIGYKAIDGGVTRDIASTDTCFANKHADFDTWFSQRGGSYTNDKACPLIAAVITREVGFVVPGLPGTILRTTNGDFLHFLPRVFSAVGNICYTPSKLIEYTDFATSACVLAAECTIFKDASGKPVPYCYDTNVLEGSVAYESLRPDTRYVLMDGSIIQFPNTYLEGSVRVVTTFDSEYCRHGTCERSEAGVCVSTSGRWVLNNDYYRSLPGVFCGVDAVNLLTNMFTPLIQPIGALDISASIVAGGIVAIVVTCLAYYFMRFRRAFGEYSHVVAFNTLLFLMSFTVLCLTPVYSFLPGVYSVIYLYLTFYLTNDVSFLAHIQWMVMFTPLVPFWITIAYIICISTKHFYWFFSNYLKRRVVFNGVSFSTFEEAALCTFLLNKEMYLKLRSDVLLPLTQYNRYLALYNKYKYFSGAMDTTSYREAACCHLAKALNDFSNSGSDVLYQPPQTSITSAVLQ
Participates in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication [https://swissmodel.expasy.org/repository/species/2697049, https://jvi.asm.org/content/83/19/10314]
3D-Structures
No known experimental structures yet
nsp5 (306 amino acids):
Residue 3264-3569 in orf1ab polyprotein SGFRKMAFPSGKVEGCMVQVTCGTTTLNGLWLDDVVYCPRHVICTSEDMLNPNYEDLLIRKSNHNFLVQAGNVQLRVIGHSMQNCVLKLKVDTANPKTPKYKFVRIQPGQTFSVLACYNGSPSGVYQCAMRPNFTIKGSFLNGSCGSVGFNIDYDCVSFCYMHHMELPTGVHAGTDLEGNFYGPFVDRQTAQAAGTDTTITVNVLAWLYAAVINGDRWFLNRFTTTLNDFNLVAMKYNYEPLTQDHVDILGPLSAQTGIAVLDMCASLKELLQNGMNGRTILGSALLEDEFTPFDVVRQCSGVTFQ
Alternative names: 3C-like proteinase, 3CLpro, Mpro (that’s the protease a lot of people are after right now)
Cleaves the C-terminus of replicase polyprotein at 11 sites. Recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN]. Also able to bind an ADP-ribose-1'- phosphate (ADRP).
3D-Structures:
a lot of structures are available with different fragments bound into the active site
nsp6 (290 amino acids):
Residue 3570-3859 in orf1ab polyprotein SAVKRTIKGTHHWLLLTILTSLLVLVQSTQWSLFFFLYENAFLPFAMGIIAMSAFAMMFVKHKHAFLCLFLLPSLATVAYFNMVYMPASWVMRIMTWLDMVDTSLSGFKLKDCVMYASAVVLLILMTARTVYDDGARRVWTLMNVLTLVYKVYYGNALDQAISMWALIISVTSNYSGVVTTVMFLARGIVFMCVEYCPIFFITGNTLQCIMLVYCFLGYFCTCYFGLFCLLNRYFRLTLGVYDYLVSTQEFRYMNSQGLLPPKNSIDAFKLNIKLLGVGGKPCIKVATVQ
Plays a role in the initial induction of autophagosomes from host reticulum endoplasmic. Later, limits the expansion of these phagosomes that are no longer able to deliver viral components to lysosomes.
3D-Structures:
no known experimental structures
nsp7 (63 amino acids):
Residue 3860-3942 in orf1ab polyprotein
SKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDILLAKDTTEAFEKMVSLLSVLLSMQGAVDINKLCEEMLDNRATLQ
Cofactor with nsp8 and nsp12
Forms a hexadecamer with nsp8 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers. May be required to activate RNA-synthesizing activity of Pol [https://swissmodel.expasy.org/repository/species/2697049 , https://jvi.asm.org/content/83/19/10314]
3D-Structures:
no known experimental structures
nsp8 (198 amino acids):
Residue 3943-4140 in orf1ab polyprotein AIASEFSSLPSYAAFATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLEKMADQAMTQMYKQARSEDKRAKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNIIPLTTAAKLMVVIPDYNTYKNTCDGTTFTYASALWEIQQVVDADSKIVQLSEISMDNSPNLAWPLIVTALRANSAVKLQ
Cofactor with nsp7 and nsp12, primase
Forms a hexadecamer with nsp7 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers. May be required to activate RNA-synthesizing activity of Pol [https://swissmodel.expasy.org/repository/species/2697049 , https://jvi.asm.org/content/83/19/10314]
3D-Structures:
no known experimental structures
nsp9 (113 amino acids):
Residue 4141-4253 in orf1ab polyprotein NNELSPVALRQMSCAAGTTQTACTDDNALAYYNTTKGGRFVLALLSDLQDLKWARFPKSDGTGTIYTELEPPCRFVTDTPKGPKVKYLYFIKGLNNLNRGMVLGSLAATVRLQ
May participate in viral replication by acting as a ssRNA-binding protein.
Dimerization [https://swissmodel.expasy.org/repository/species/2697049 , https://jvi.asm.org/content/83/19/10314]
3D-Structures:
experimental structures are known for the full sequence
nsp10 (139 amino acids):
Residue 4254-4392 in orf1ab polyprotein AGNATEVPANSTVLSFCAFAVDAAKAYKDYLASGGQPITNCVKMLCTHTGTGQAITVTPEANMDQESFGGASCCLYCRCHIDHPNPKGFCDLKGKYVQIPTTCANDPVGFTLKNTVCTVCGMWKGYGCSCDQLREPMLQ
Plays a pivotal role in viral transcription by stimulating both nsp14 3'-5' exoribonuclease and nsp16 2'-O-methyltransferase activities. Therefore plays an essential role in viral mRNAs cap methylation [https://swissmodel.expasy.org/repository/species/2697049]
Cofactor for nsp16 and nsp14, forms heterodimer with both and stimulates ExoN and 2-O-MT activity [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4369385/]
3D-Structures:
experimental structures are available of the full domain
nsp11 (13 amino acids):
Residue 4393-4405 in orf1a polyprotein
Becomes nsp12 in orf1ab polyprotein
nsp12 (932 amino acids):
Residue 4393-5324 in orf1ab polyprotein SADAQSFLNRVCGVSAARLTPCGTGTSTDVVYRAFDIYNDKVAGFAKFLKTNCCRFQEKDEDDNLIDSYFVVKRHTFSNYQHEETIYNLLKDCPAVAKHDFFKFRIDGDMVPHISRQRLTKYTMADLVYALRHFDEGNCDTLKEILVTYNCCDDDYFNKKDWYDFVENPDILRVYANLGERVRQALLKTVQFCDAMRNAGIVGVLTLDNQDLNGNWYDFGDFIQTTPGSGVPVVDSYYSLLMPILTLTRALTAESHVDTDLTKPYIKWDLLKYDFTEERLKLFDRYFKYWDQTYHPNCVNCLDDRCILHCANFNVLFSTVFPPTSFGPLVRKIFVDGVPFVVSTGYHFRELGVVHNQDVNLHSSRLSFKELLVYAADPAMHAASGNLLLDKRTTCFSVAALTNNVAFQTVKPGNFNKDFYDFAVSKGFFKEGSSVELKHFFFAQDGNAAISDYDYYRYNLPTMCDIRQLLFVVEVVDKYFDCYDGGCINANQVIVNNLDKSAGFPFNKWGKARLYYDSMSYEDQDALFAYTKRNVIPTITQMNLKYAISAKNRARTVAGVSICSTMTNRQFHQKLLKSIAATRGATVVIGTSKFYGGWHNMLKTVYSDVENPHLMGWDYPKCDRAMPNMLRIMASLVLARKHTTCCSLSHRFYRLANECAQVLSEMVMCGGSLYVKPGGTSSGDATTAYANSVFNICQAVTANVNALLSTDGNKIADKYVRNLQHRLYECLYRNRDVDTDFVNEFYAYLRKHFSMMILSDDAVVCFNSTYASQGLVASIKNFKSVLYYQNNVFMSEAKCWTETDLTKGPHEFCSQHTMLVKQGDDYVYLPYPDPSRILGAGCFVDDIVKTDGTLMIERFVSLAIDAYPLTKHPNQEYADVFHLYLQYIRKLHDELTGHMLDMYSVMLTNDNTSRYWEPEFYEAMYTPHTVLQ
Alternative names:
RNA directed RNA polymerase
Pol/RdRp
Responsible for replication and transcription of the viral RNA genome. [https://swissmodel.expasy.org/repository/species/2697049]
3D-Structures:
no known experimental structure
nsp13 (601 amino acids):
Residue 5325-5925 in orf1ab polyprotein AVGACVLCNSQTSLRCGACIRRPFLCCKCCYDHVISTSHKLVLSVNPYVCNAPGCDVTDVTQLYLGGMSYYCKSHKPPISFPLCANGQVFGLYKNTCVGSDNVTDFNAIATCDWTNAGDYILANTCTERLKLFAAETLKATEETFKLSYGIATVREVLSDRELHLSWEVGKPRPPLNRNYVFTGYRVTKNSKVQIGEYTFEKGDYGDAVVYRGTTTYKLNVGDYFVLTSHTVMPLSAPTLVPQEHYVRITGLYPTLNISDEFSSNVANYQKVGMQKYSTLQGPPGTGKSHFAIGLALYYPSARIVYTACSHAAVDALCEKALKYLPIDKCSRIIPARARVECFDKFKVNSTLEQYVFCTVNALPETTADIVVFDEISMATNYDLSVVNARLRAKHYVYIGDPAQLPAPRTLLTKGTLEPEYFNSVCRLMKTIGPDMFLGTCRRCPAEIVDTVSALVYDNKLKAHKDKSAQCFKMFYKGVITHDVSSAINRPQIGVVREFLTRNPAWRKAVFISPYNSQNAVASKILGLPTQTVDSSQGSEYDYVIFTQTTETAHSCNVNRFNVAITRAKVGILCIMSDRDLYDKLQFTSLEIPRRNVATLQ
Helicase (Hel)
Multi-functional protein with a zinc-binding domain in N-terminus displaying RNA and DNA duplex-unwinding activities with 5' to 3' polarity. Activity of helicase is dependent on magnesium. [https://swissmodel.expasy.org/repository/species/2697049]
3D-Structures:
no known experimental structure
nsp14 (527 amino acids):
Residue 5926-6452 in orf1ab polyprotein AENVTGLFKDCSKVITGLHPTQAPTHLSVDTKFKTEGLCVDIPGIPKDMTYRRLISMMGFKMNYQVNGYPNMFITREEAIRHVRAWIGFDVEGCHATREAVGTNLPLQLGFSTGVNLVAVPTGYVDTPNNTDFSRVSAKPPPGDQFKHLIPLMYKGLPWNVVRIKIVQMLSDTLKNLSDRVVFVLWAHGFELTSMKYFVKIGPERTCCLCDRRATCFSTASDTYACWHHSIGFDYVYNPFMIDVQQWGFTGNLQSNHDLYCQVHGNAHVASCDAIMTRCLAVHECFVKRVDWTIEYPIIGDELKINAACRKVQHMVVKAALLADKFPVLHDIGNPKAIKCVPQADVEWKFYDAQPCSDKAYKIEELFYSYATHSDKFTDGVCLFWNCNVDRYPANSIVCRFDTRVLSNLNLPGCDGGSLYVNKHAFHTPAFDKSAFVNLKQLPFFYYSDSPCESHGKQVVSDIDYVPLKSATCITRCNLGGAVCRHHANEYRLYLDAYNMMISAGFSLWVYKQFDTYNLWNTFTRLQ
Alternative names:
Guanine-N7 methyltransferase
ExoN/nsp14
Enzyme possessing two different activities: an exoribonuclease activity acting on both ssRNA and dsRNA in a 3' to 5' direction and a N7-guanine methyltransferase activity.
3D-Structures:
no known experimental structure
nsp15 (346 amino acids):
Residue 6453-6798 in orf1ab polyprotein SLENVAFNVVNKGHFDGQQGEVPVSIINNTVYTKVDGVDVELFENKTTLPVNVAFELWAKRNIKPVPEVKILNNLGVDIAANTVIWDYKRDAPAHISTIGVCSMTDIAKKPTETICAPLTVFFDGRVDGQVDLFRNARNGVLITEGSVKGLQPSVGPKQASLNGVTLIGEAVKTQFNYYKKVDGVVQQLPETYFTQSRNLQEFKPRSQMEIDFLELAMDEFIERYKLEGYAFEHIVYGDFSHSQLGGLHLLIGLAKRFKESPFELEDFIPMDSTVKNYFITDAQTGSSKCVCSVIDLLLDDFVEIIKSQDLSVVSKVVKVTIDYTEISFMLWCKDGHVETFYPKLQ
Alternative names:
Uridylate-specific endoribonuclease
NendoU/nsp15
Mn(2+)-dependent, uridylate-specific enzyme, which leaves 2'-3'-cyclic phosphates 5' to the cleaved bond.[https://swissmodel.expasy.org/repository/species/2697049]
3D-Structures:
Experimental structures available
nsp16 (298 amino acids):
Residue 6799-7096 in orf1ab polyprotein SSQAWQPGVAMPNLYKMQRMLLEKCDLQNYGDSATLPKGIMMNVAKYTQLCQYLNTLTLAVPYNMRVIHFGAGSDKGVAPGTAVLRQWLPTGTLLVDSDLNDFVSDADSTLIGDCATVHTANKWDLIISDMYDPKTKNVTKENDSKEGFFTYICGFIQQKLALGGSVAIKITEHSWNADLYKLMGHFAWWTAFVTNVNASSSEAFLIGCNYLGKPREQIDGYVMHANYIFWRNTNPIQLSSYSLFDMSKFPLKLRGTAVMSLKEGQINDMILSLLSKGRLIIRENNRVVISSDVLVNN
Alternative names:
2'-O-ribose methyltransferase
Methyltransferase that mediates mRNA cap 2'-O-ribose methylation to the 5'-cap structure of viral mRNAs. N7-methyl guanosine cap is a prerequisite for binding of nsp16. Therefore plays an essential role in viral mRNAs cap methylation which is essential to evade immune system.
2′‐O‐MTase; avoiding MDA5 recognition, negatively regulating innate immunity
3D-Structures:
experimental structures are available
Functional annotations of structural proteins
Spike glycoprotein MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDDSEPVLKGVKLHYT
Spike protein S1 (residue 14-685): attaches the virion to the cell membrane by interacting with host receptor, initiating the infection. Binding to human ACE2 and CLEC4M/DC-SIGNR receptors and internalization of the virus into the endosomes of the host cell induces conformational changes in the S glycoprotein. Proteolysis by cathepsin CTSL may unmask the fusion peptide of S2 and activate membranes fusion within endosomes.
Spike protein S2 (residue 686-1273): mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. Under the current model, the protein has at least three conformational states: pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During viral and target cell membrane fusion, the coiled coil regions (heptad repeats) assume a trimer-of-hairpins structure, positioning the fusion peptide in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive apposition and subsequent fusion of viral and target cell membranes.
Spike protein S2' (residue 816-1273): acts as a viral fusion peptide which is unmasked following S2 cleavage occurring upon virus endocytosis.
https://swissmodel.expasy.org/repository/species/2697049, https://zhanglab.ccmb.med.umich.edu/C-I-TASSER/2019-nCov/, https://www.ncbi.nlm.nih.gov/protein/1791269090
3D-Structures:
experimental structures are available
Protein 3a MDLFMRIFTIGTVTLKQGEIKDATPSDFVRATATIPIQASLPFGWLIVGVALLAVFQSASKIITLKKRWQLALSKGVHFVCNLLLLFVTVYSHLLLVAAGLEAPFLYLYALVYFLQSINFVRIIMRLWLCWKCRSKNPLLYDANYFLCWHTNCYDYCIPYNSVTSSIVITSGDGTTSPISEHDYQIGGYTEKWESGVKDCVVLHSYFTSDYYQLYSTQLSTDTGVEHVTFFIYNKIVDEPEEHVQIHTIDGSSGVVNPVMEPIYDEPTTTTSVPL
Forms homotetrameric potassium sensitive ion channels (viroporin) and may modulate virus release. Up-regulates expression of fibrinogen subunits FGA, FGB and FGG in host lung epithelial cells. Induces apoptosis in cell culture. Downregulates the type 1 interferon receptor by inducing serine phosphorylation within the IFN alpha- receptor subunit 1 (IFNAR1) degradation motif and increasing IFNAR1 ubiquitination.
3D-Structures:
no known experimental structure
Envelope small protein (E protein) MYSFVSEETGTLIVNSVLLFLAFVVFLLVTLAILTALRLCAYCCNIVNVSLVKPSFYVYSRVKNLNSSRVPDLLV
Plays a central role in virus morphogenesis and assembly. Acts as a viroporin and self-assembles in host membranes forming pentameric protein-lipid pores that allow ion transport. Also plays a role in the induction of apoptosis. Activates the host NLRP3 inflammasome, leading to IL-1beta overproduction.
3D-Structures:
no known experimental structure
Membrane protein (M protein) MADSNGTITVEELKKLLEQWNLVIGFLFLTWICLLQFAYANRNRFLYIIKLIFLWLLWPVTLACFVLAAVYRINWITGGIAIAMACLVGLMWLSYFIASFRLFARTRSMWSFNPETNILLNVPLHGTILTRPLLESELVIGAVILRGHLRIAGHHLGRCDIKDLPKEITVATSRTLSYYKLGASQRVAGDSGFAAYSRYRIGNYKLNTDHSSSSDNIALLVQ
Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral protein
3D-Structures:
no known experimental structure
ns6 (ORF6):
MFHLVDFQVTIAEILLIIMRTFKVSIWNLDYIINLIIKNLSKSLTENKYSQLDEEQPMEIDCould be a determinant of virus virulence, since, when expressed in an otherwise attenuated JHM strain of murine coronavirus, it can dramatically increase the lethality of the latter. Seems to stimulate cellular DNA synthesis in vitro.
3D-Structures:
no known experimental structure
Protein 7a (ns7): MKIILFLALITLATCELYHYQECVRGTTVLLKEPCSSGTYEGNSPFHPLADNKFALTCFSTQFAFACPDGVKHVYQLRARSVSPKLFIRQEEVQELYSPIFLIVAAIVFITLCFTLKRKTE
Non-structural protein which is dispensable for virus replication in cell culture. Suppression of host tetherin activity.
3D-Structures:
no known experimental structure
Protein 7b
NA
ns 8 (ORF8) MKFLVFLGIITTVAAFHQECSLQSCTQHQPYVVDDPCPIHFYSKWYIRVGARKSAPLIELCVDEAGSKSPIQYIDIGNYTVSCLPFTINCQEPKLGSLVVRCSFYEDFLEYHDVRVVLDFI
May play a role in host-virus interaction.
3D-Structures:
no known experimental structure
N protein MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSKQRRPQGLPNNTASWFTALTQHGKEDLKFPRGQGVPINTNSSPDDQIGYYRRATRRIRGGDGKMKDLSPRWYFYYLGTGPEAGLPYGANKDGIIWVATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGSRGGSQASSRSSSRSRNSSRNSTPGSSRGTSPARMAGNGGDAALALLLLDRLNQLESKMSGKGQQQQGQTVTKKSAAEASKKPRQKRTATKAYNVTQAFGRRGPEQTQGNFGDQELIRQGTDYKHWPQIAQFAPSASAFFGMSRIGMEVTPSGTWLTYTGAIKLDDKDPNFKDQVILLNKHIDAYKTFPPTEPKKDKKKKADETQALPQRQKKQQTVTLLPAADLDDFSKQLQQSMSSADSTQA
Packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP) and plays a fundamental role during virion assembly through its interactions with the viral genome and membrane protein M. Plays an important role in enhancing the efficiency of subgenomic viral RNA transcription as well as viral replication.
3D-Structures:
one domain is resolved experimentally
ORF10 protein
MGYINVFAFPFTIYSLLLCRMNSRNYIAQVDVVNFNLTDubious/uncertain protein. It is currently unclear whether this region translates into a functional protein. The respective region in related beta-coronaviruses, such as SARS-CoV, does not correspond to translated peptide.
3D-Structures:
no known structures
Therapeutic options
Here I’ll mainly focus on non-preventive measures outlined in Chen et al. Let’s make it short, this quote from that paper states rather clearly where we stand today:
“After SARS and MERS epidemics, great efforts have been devoted to development of new antivirals targeting CoVs proteases, polymerases, MTases, and entry proteins, however, none of them has been shown to be efficacious in clinical trials. Plasma and antibodies obtained from the convalescent patients have been proposed for use in treatment.”
Some of you might disagree in light of some recent trial results, but the fact is that there is no efficient treatment against Covid-19 known as of today. It will be very interesting to see whether the global effort to find a treatment quickly will change the fact that previous efforts on the previous SARS and MERS outbreaks failed. Likely emerging coronaviruses have now the attention they should have gotten during these last outbreaks.
During the next days we will analyse in more depth each constituent of the virus, treatments tried during MERS and SARS outbreaks and potential hit ID discovery strategies, modelling and screening efforts etc.
Another short review that appeared in NRDD recently lists all currently ongoing tests with existing antivirals.
The following proteins have been mentioned as of high interest (and priority) for drug discovery programs for antivirals because of the obvious important function for the virus:
Mpro (the main protease, nsp5) (structures available)
papain-like protease (nsp3) (structures available)
helicase (nsp13) (good models available)
RNA dependent RNA polymerase (nsp12) (good models available)
spike glycoprotein (structures available)
A very interesting paper (yet to be validated, as a lot of them right now) came out during the last week-end here. It states a few known interactions from previous SARS-Cov and MERS studies, but it also adds a lot of other potentially interesting interactions. It has been performed on a limited set of host proteins, so the view is by far not complete and should be thoroughly completed with a proper literature search.
Interaction map from https://www.biorxiv.org/content/10.1101/2020.03.22.002386v3
There are also some interesting things published by the Korkin lab here. I’ll add them as soon I as start to analyse the relevant proteins in the upcoming posts.
Some antivirals currently under investigation:
This list is more less long according to the sources and changing a lot as everybody is trying to repurpose drugs right now. So a few of the examples tested in humans.
Favipiravir:
inhibitor of the RNA dependent RNA polymerase (approved influenza treatment)
EC50= 61.88µM SARS-Cov-2 [https://doi.org/10.1038/s41422-020-0282-0, https://www.theguardian.com/world/2020/mar/18/japanese-flu-drug-clearly-effective-in-treating-coronavirus-says-china]
CID 492405 ZCGNOVWYSGBHAU-UHFFFAOYSA-N
Ribavirin:
severe side-effects at high doses
guanine derivative approved against HCV
mode of action in SARS-Cov-2 not clear (potential 2'-O-ribose methyltransferase inhibitor?)
CID 37542 IWUCXVSUMQZMFG-AFCXAGJDSA-N
Remdesivir:
EC50=0.77µM SARS-Cov-2
Results so far: https://www.statnews.com/2020/03/16/remdesivir-surges-ahead-against-coronavirus/
CID 121304016 RWWYLEGWBNMMLJ-YSOARWBDSA-N
Galidesivir:
RNA dependent RNA polymerase inhibitor
CID 10445549 AMFDITJFBUXZQN-KUBHLMPHSA-N
Griffithsin:
targeting spike glycoprotein
binds to oligosaccharides on surface of various proteins
Chloroquine (attracted a lot attention lately):
EC50 = 1.13 μM SARS-Cov-2
Chloroquine passively diffuses through cell membranes and into endosomes, lysosomes, and Golgi vesicles; where it becomes protonated, trapping the chloroquine in the organelle and raising the surrounding pH.10,13 The raised pH in endosomes, prevent virus particles from utilizing their activity for fusion and entry into the cell.14
Chloroquine does not affect the level of ACE2 expression on cell surfaces, but inhibits terminal glycosylation of ACE2, the receptor that SARS-CoV and SARS-CoV-2 target for cell entry.13,14 ACE2 that is not in the glycosylated state may less efficiently interact with the SARS-CoV-2 spike protein, further inhibiting viral entry.14 [https://www.drugbank.ca/drugs/DB00608]
Nitazoxanide:
EC50 = 2.12 μM SARS-Cov-2
mode of action not known?
CID 41684 YQNQNVDNTFHQSW-UHFFFAOYSA-N
Disulfiram:
active against MERS & SARS-Cov
CID 3117 AUZONCFQVSMFAP-UHFFFAOYSA-N
Lopinavir / Ritonavir:
HIV protease inhibitor
no action: https://www.nejm.org/doi/full/10.1056/NEJMoa2001282?query=featured_home
CID 92727 KJHKTHWMRKYKJE-SUGCFTRWSA-N
CID 392622 NCDNCNXCDXHOMX-XGKFQTDJSA-N
Structural Resources to follow
Excellent article to read in the NYtimes